Portraits of breast cancer progression
- PMID: 17683614
- PMCID: PMC1978212
- DOI: 10.1186/1471-2105-8-291
Portraits of breast cancer progression
Abstract
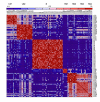
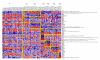
Background: Clustering analysis of microarray data is often criticized for giving ambiguous results because of sensitivity to data perturbation or clustering techniques used. In this paper, we describe a new method based on principal component analysis and ensemble consensus clustering that avoids these problems.
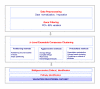
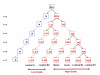
Results: We illustrate the method on a public microarray dataset from 36 breast cancer patients of whom 31 were diagnosed with at least two of three pathological stages of disease (atypical ductal hyperplasia (ADH), ductal carcinoma in situ (DCIS) and invasive ductal carcinoma (IDC). Our method identifies an optimum set of genes and divides the samples into stable clusters which correlate with clinical classification into Luminal, Basal-like and Her2+ subtypes. Our analysis reveals a hierarchical portrait of breast cancer progression and identifies genes and pathways for each stage, grade and subtype. An intriguing observation is that the disease phenotype is distinguishable in ADH and progresses along distinct pathways for each subtype. The genetic signature for disease heterogeneity across subtypes is greater than the heterogeneity of progression from DCIS to IDC within a subtype, suggesting that the disease subtypes have distinct progression pathways. Our method identifies six disease subtype and one normal clusters. The first split separates the normal samples from the cancer samples. Next, the cancer cluster splits into low grade (pathological grades 1 and 2) and high grade (pathological grades 2 and 3) while the normal cluster is unchanged. Further, the low grade cluster splits into two subclusters and the high grade cluster into four. The final six disease clusters are mapped into one Luminal A, three Luminal B, one Basal-like and one Her2+.
Conclusion: We confirm that the cancer phenotype can be identified in early stage because the genes altered in this stage progressively alter further as the disease progresses through DCIS into IDC. We identify six subtypes of disease which have distinct genetic signatures and remain separated in the clustering hierarchy. Our findings suggest that the heterogeneity of disease across subtypes is higher than the heterogeneity of the disease progression within a subtype, indicating that the subtypes are in fact distinct diseases.
Figures



References
-
- Gruvberger S, Ringner M, Chen Y, Panavally S, Saal LH, Borg A, Ferno M, Peterson C, Meltzer PS. Estrogen receptor status in breast cancer is associated with remarkably distinct gene expression patterns. Cancer Res. 2001;61:5979–5984. - PubMed
-
- Mauriac L. Aromatase inhibitors: Effective endocrine therapy in the early adjuvant setting for postmenopausal women with hormone-responsive breast cancer. Best Pract Res Clin Endocrinol Metab. 2006;20:S15–29.
-
- Morris SR, Carey LA. Molecular profiling in breast cancer. Rev Endocr Metab Disord. 2007 - PubMed
-
- Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

