SNPs in Multi-species Conserved Sequences (MCS) as useful markers in association studies: a practical approach
- PMID: 17683615
- PMCID: PMC1959193
- DOI: 10.1186/1471-2164-8-266
SNPs in Multi-species Conserved Sequences (MCS) as useful markers in association studies: a practical approach
Abstract
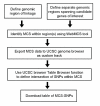
Background: Although genes play a key role in many complex diseases, the specific genes involved in most complex diseases remain largely unidentified. Their discovery will hinge on the identification of key sequence variants that are conclusively associated with disease. While much attention has been focused on variants in protein-coding DNA, variants in noncoding regions may also play many important roles in complex disease by altering gene regulation. Since the vast majority of noncoding genomic sequence is of unknown function, this increases the challenge of identifying "functional" variants that cause disease. However, evolutionary conservation can be used as a guide to indicate regions of noncoding or coding DNA that are likely to have biological function, and thus may be more likely to harbor SNP variants with functional consequences. To help bias marker selection in favor of such variants, we devised a process that prioritizes annotated SNPs for genotyping studies based on their location within Multi-species Conserved Sequences (MCSs) and used this process to select SNPs in a region of linkage to a complex disease. This allowed us to evaluate the utility of the chosen SNPs for further association studies. Previously, a region of chromosome 1q43 was linked to Multiple Sclerosis (MS) in a genome-wide screen. We chose annotated SNPs in the region based on location within MCSs (termed MCS-SNPs). We then obtained genotypes for 478 MCS-SNPs in 989 individuals from MS families.
Results: Analysis of our MCS-SNP genotypes from the 1q43 region and comparison to HapMap data confirmed that annotated SNPs in MCS regions are frequently polymorphic and show subtle signatures of selective pressure, consistent with previous reports of genome-wide variation in conserved regions. We also present an online tool that allows MCS data to be directly exported to the UCSC genome browser so that MCS-SNPs can be easily identified within genomic regions of interest.
Conclusion: Our results showed that MCS can easily be used to prioritize markers for follow-up and candidate gene association studies. We believe that this novel approach demonstrates a paradigm for expediting the search for genes contributing to complex diseases.
Figures
Similar articles
-
Follow-up examination of linkage and association to chromosome 1q43 in multiple sclerosis.Genes Immun. 2009 Oct;10(7):624-30. doi: 10.1038/gene.2009.53. Epub 2009 Jul 23. Genes Immun. 2009. PMID: 19626040 Free PMC article.
-
Linkage mapping bovine EST-based SNP.BMC Genomics. 2005 May 19;6:74. doi: 10.1186/1471-2164-6-74. BMC Genomics. 2005. PMID: 15943875 Free PMC article.
-
Selecting SNPs for association studies based on population frequencies: a novel interactive tool and its application to polygenic diseases.In Silico Biol. 2004;4(4):417-27. In Silico Biol. 2004. PMID: 15506992
-
From genetic associations to functional studies in multiple sclerosis.Eur J Neurol. 2016 May;23(5):847-53. doi: 10.1111/ene.12981. Epub 2016 Mar 7. Eur J Neurol. 2016. PMID: 26948534 Review.
-
Tag SNP selection for association studies.Genet Epidemiol. 2004 Dec;27(4):365-74. doi: 10.1002/gepi.20028. Genet Epidemiol. 2004. PMID: 15372618 Review.
Cited by
-
Strategies for the identification of loci responsible for the pathogenesis of multiple sclerosis.Cell Mol Biol Lett. 2008;13(4):656-66. doi: 10.2478/s11658-008-0030-9. Epub 2008 Aug 23. Cell Mol Biol Lett. 2008. PMID: 18726192 Free PMC article. Review.
-
SNP eQTL status and eQTL density in the adjacent region of the SNP are associated with its statistical significance in GWA studies.BMC Genet. 2019 Nov 12;20(1):85. doi: 10.1186/s12863-019-0786-0. BMC Genet. 2019. PMID: 31718536 Free PMC article.
-
The top five "game changers" in vaccinology: toward rational and directed vaccine development.OMICS. 2011 Sep;15(9):533-7. doi: 10.1089/omi.2011.0012. Epub 2011 Aug 4. OMICS. 2011. PMID: 21815811 Free PMC article. Review.
-
Population differences in transcript-regulator expression quantitative trait loci.PLoS One. 2012;7(3):e34286. doi: 10.1371/journal.pone.0034286. Epub 2012 Mar 27. PLoS One. 2012. PMID: 22479588 Free PMC article.
-
Multiple hits for the association of uterine fibroids on human chromosome 1q43.PLoS One. 2013;8(3):e58399. doi: 10.1371/journal.pone.0058399. Epub 2013 Mar 14. PLoS One. 2013. PMID: 23555580 Free PMC article. Clinical Trial.
References
-
- Hauser MA, Li YJ, Takeuchi S, Walters R, Noureddine M, Maready M, Darden T, Hulette C, Martin E, Hauser E, et al. Genomic convergence: identifying candidate genes for Parkinson's disease by combining serial analysis of gene expression and genetic linkage. Hum Mol Genet. 2003;12:671–7. doi: 10.1093/hmg/12.6.671. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

