Analysis of the Rana catesbeiana tadpole tail fin proteome and phosphoproteome during T3-induced apoptosis: identification of a novel type I keratin
- PMID: 17683616
- PMCID: PMC2025591
- DOI: 10.1186/1471-213X-7-94
Analysis of the Rana catesbeiana tadpole tail fin proteome and phosphoproteome during T3-induced apoptosis: identification of a novel type I keratin
Abstract
Background: Thyroid hormones (THs) are vital in the maintenance of homeostasis and in the control of development. One postembryonic developmental process that is principally regulated by THs is amphibian metamorphosis. This process has been intensively studied at the genomic level yet very little information at the proteomic level exists. In addition, there is increasing evidence that changes in the phosphoproteome influence TH action.
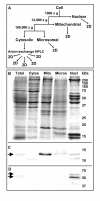
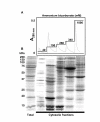
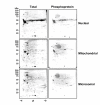
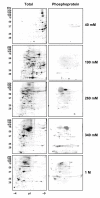
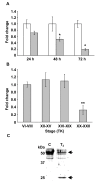
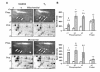
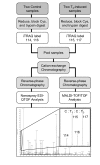
Results: Here we identify components of the proteome and phosphoproteome in the tail fin that changed within 48 h of exposure of premetamorphic Rana catesbeiana tadpoles to 10 nM 3,5,3'-triiodothyronine (T3). To this end, we developed a cell and protein fractionation method combined with two-dimensional gel electrophoresis and phosphoprotein-specific staining. Altered proteins were identified using mass spectrometry (MS). We identified and cloned a novel Rana larval type I keratin, RLK I, which may be a target for caspase-mediated proteolysis upon exposure to T3. In addition, the RLK I transcript is reduced during T3-induced and natural metamorphosis which is consistent with a larval keratin. Furthermore, GILT, a protein involved in the immune system, is changed in phosphorylation state which is linked to its activation. Using a complementary MS technique for the analysis of differentially-expressed proteins, isobaric tags for relative and absolute quantitation (iTRAQ) revealed 15 additional proteins whose levels were altered upon T3 treatment. The success of identifying proteins whose levels changed upon T3 treatment with iTRAQ was enhanced through de novo sequencing of MS data and homology database searching. These proteins are involved in apoptosis, extracellular matrix structure, immune system, metabolism, mechanical function, and oxygen transport.
Conclusion: We have demonstrated the ability to derive proteomics-based information from a model species for postembryonic development for which no genome information is currently available. The present study identifies proteins whose levels and/or phosphorylation states are altered within 48 h of the induction of tadpole tail regression prior to overt remodeling of the tail. In particular, we have identified a novel keratin that is a target for T3-mediated changes in the tail that can serve as an indicator of early response to this hormone.
Figures



Similar articles
-
Novel Rana keratin genes and their expression during larval to adult epidermal conversion in bullfrog tadpoles.Differentiation. 2001 Aug;68(1):44-54. doi: 10.1046/j.1432-0436.2001.068001044.x. Differentiation. 2001. PMID: 11683492
-
The bactericidal agent triclosan modulates thyroid hormone-associated gene expression and disrupts postembryonic anuran development.Aquat Toxicol. 2006 Dec 1;80(3):217-27. doi: 10.1016/j.aquatox.2006.08.010. Epub 2006 Sep 29. Aquat Toxicol. 2006. PMID: 17011055
-
Behavioral and molecular analyses of olfaction-mediated avoidance responses of Rana (Lithobates) catesbeiana tadpoles: Sensitivity to thyroid hormones, estrogen, and treated municipal wastewater effluent.Horm Behav. 2018 May;101:85-93. doi: 10.1016/j.yhbeh.2017.09.016. Epub 2017 Oct 4. Horm Behav. 2018. PMID: 28964734
-
[Induction of cell differentiation and programmed cell death in amphibian metamorphosis].Hum Cell. 1997 Sep;10(3):167-74. Hum Cell. 1997. PMID: 9436036 Review. Japanese.
-
Postembryonic expression of the myosin heavy chain genes in the limb, tail, and heart muscles of metamorphosing amphibian tadpoles.Microsc Res Tech. 2000 Sep 15;50(6):458-72. doi: 10.1002/1097-0029(20000915)50:6<458::AID-JEMT4>3.0.CO;2-V. Microsc Res Tech. 2000. PMID: 10998636 Review.
Cited by
-
Influence of Nitrate and Nitrite on Thyroid Hormone Responsive and Stress-Associated Gene Expression in Cultured Rana catesbeiana Tadpole Tail Fin Tissue.Front Genet. 2012 Apr 4;3:51. doi: 10.3389/fgene.2012.00051. eCollection 2012. Front Genet. 2012. PMID: 22493607 Free PMC article.
-
Integrative proteome and metabolome analyses reveal molecular basis of the tail resorption during the metamorphic climax of Nanorana pleskei.Front Cell Dev Biol. 2024 Aug 19;12:1431173. doi: 10.3389/fcell.2024.1431173. eCollection 2024. Front Cell Dev Biol. 2024. PMID: 39224435 Free PMC article.
-
Evaluation of the effects of titanium dioxide nanoparticles on cultured Rana catesbeiana tailfin tissue.Front Genet. 2013 Nov 21;4:251. doi: 10.3389/fgene.2013.00251. eCollection 2013. Front Genet. 2013. PMID: 24312126 Free PMC article.
-
Apoptosis in amphibian organs during metamorphosis.Apoptosis. 2010 Mar;15(3):350-64. doi: 10.1007/s10495-009-0422-y. Apoptosis. 2010. PMID: 20238476 Free PMC article. Review.
-
DIGE and iTRAQ as biomarker discovery tools in aquatic toxicology.Ecotoxicol Environ Saf. 2012 Feb;76(2):3-10. doi: 10.1016/j.ecoenv.2011.09.020. Epub 2011 Nov 5. Ecotoxicol Environ Saf. 2012. PMID: 22056798 Free PMC article. Review.
References
-
- Shi YB. Amphibian Metamorphosis: from morphology to molecular biology. New York, Wiley-Liss; 2000.
-
- Regard E, Taurog A, Nakashima T. Plasma thyroxine and triiodothyronine levels in spontaneously metamorphosing Rana catesbeiana tadpoles and in adult anuran amphibia. Endocrinology. 1978;102:674–684. - PubMed
-
- Wang Z, Brown DD. Thyroid hormone-induced gene expression program for amphibian tail resorption. J Biol Chem. 1993;268:16270–16278. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

