The GlycanBuilder: a fast, intuitive and flexible software tool for building and displaying glycan structures
- PMID: 17683623
- PMCID: PMC1994674
- DOI: 10.1186/1751-0473-2-3
The GlycanBuilder: a fast, intuitive and flexible software tool for building and displaying glycan structures
Abstract
Background: Carbohydrates play a critical role in human diseases and their potential utility as biomarkers for pathological conditions is a major driver for characterization of the glycome. However, the additional complexity of glycans compared to proteins and nucleic acids has slowed the advancement of glycomics in comparison to genomics and proteomics. The branched nature of carbohydrates, the great diversity of their constituents and the numerous alternative symbolic notations, make the input and display of glycans not as straightforward as for example the amino-acid sequence of a protein. Every glycoinformatic tool providing a user interface would benefit from a fast, intuitive, appealing mechanism for input and output of glycan structures in a computer readable format.
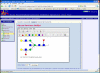
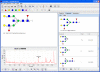
Results: A software tool for building and displaying glycan structures using a chosen symbolic notation is described here. The "GlycanBuilder" uses an automatic rendering algorithm to draw the saccharide symbols and to place them on the drawing board. The information about the symbolic notation is derived from a configurable graphical model as a set of rules governing the aspect and placement of residues and linkages. The algorithm is able to represent a structure using only few traversals of the tree and is inherently fast. The tool uses an XML format for import and export of encoded structures.
Conclusion: The rendering algorithm described here is able to produce high-quality representations of glycan structures in a chosen symbolic notation. The automated rendering process enables the "GlycanBuilder" to be used both as a user-independent component for displaying glycans and as an easy-to-use drawing tool. The "GlycanBuilder" can be integrated in web pages as a Java applet for the visual editing of glycans. The same component is available as a web service to render an encoded structure into a graphical format. Finally, the "GlycanBuilder" can be integrated into other applications to create intuitive and appealing user interfaces: an example is the "GlycoWorkbench", a software tool for assisted annotation of glycan mass spectra. The "GlycanBuilder" represent a flexible, reliable and efficient solution to the problem of input and output of glycan structures in any glycomic tool or database.
Figures






Similar articles
-
Implementation of GlycanBuilder to draw a wide variety of ambiguous glycans.Carbohydr Res. 2017 Jun 5;445:104-116. doi: 10.1016/j.carres.2017.04.015. Epub 2017 Apr 19. Carbohydr Res. 2017. PMID: 28525772
-
The GlycanBuilder and GlycoWorkbench glycoinformatics tools: updates and new developments.Biol Chem. 2012 Nov;393(11):1357-62. doi: 10.1515/hsz-2012-0135. Biol Chem. 2012. PMID: 23109548
-
Annotation of glycomics MS and MS/MS spectra using the GlycoWorkbench software tool.Methods Mol Biol. 2015;1273:3-15. doi: 10.1007/978-1-4939-2343-4_1. Methods Mol Biol. 2015. PMID: 25753699
-
KEGG as a glycome informatics resource.Glycobiology. 2006 May;16(5):63R-70R. doi: 10.1093/glycob/cwj010. Epub 2005 Jul 13. Glycobiology. 2006. PMID: 16014746 Review.
-
Deciphering the glycocode: the complexity and analytical challenge of glycomics.Curr Opin Chem Biol. 2007 Jun;11(3):300-5. doi: 10.1016/j.cbpa.2007.05.002. Epub 2007 May 17. Curr Opin Chem Biol. 2007. PMID: 17500024 Review.
Cited by
-
Azahar: a PyMOL plugin for construction, visualization and analysis of glycan molecules.J Comput Aided Mol Des. 2016 Aug;30(8):619-24. doi: 10.1007/s10822-016-9944-x. Epub 2016 Aug 22. J Comput Aided Mol Des. 2016. PMID: 27549814
-
Proteomics support the threespine stickleback egg coat as a protective oocyte envelope.Mol Reprod Dev. 2021 Jul;88(7):500-515. doi: 10.1002/mrd.23517. Epub 2021 Jun 20. Mol Reprod Dev. 2021. PMID: 34148267 Free PMC article.
-
Glycoinformatics Tools for Comprehensive Characterization of Glycans Enzymatically Released from Proteins.Methods Mol Biol. 2022;2370:3-23. doi: 10.1007/978-1-0716-1685-7_1. Methods Mol Biol. 2022. PMID: 34611862
-
Mass spectrometry-based analyses showing the effects of secretor and blood group status on salivary N-glycosylation.Clin Proteomics. 2015 Dec 30;12:29. doi: 10.1186/s12014-015-9100-y. eCollection 2015. Clin Proteomics. 2015. PMID: 26719750 Free PMC article.
-
Epigenetic Regulation of the Biosynthesis & Enzymatic Modification of Heparan Sulfate Proteoglycans: Implications for Tumorigenesis and Cancer Biomarkers.Int J Mol Sci. 2017 Jun 26;18(7):1361. doi: 10.3390/ijms18071361. Int J Mol Sci. 2017. PMID: 28672878 Free PMC article. Review.
References
-
- Policy Briefing 27. European Science Foundation; 2006. Structural Medicine - The Importance of Glycomics for Health and Disease.
-
- Aoki KF, von der Lieth CW, Raman R, York WS. White Paper. National Institute of Health; 2007. Urgent requirements for the development of informatics for glycomics and glycobiology.
-
- KEGG Glycan Structure Search using KCaM http://www.genome.jp/ligand/kcam/
-
- DrawRINGS, 2D Glycan Structure Drawing Tool at RINGS http://rings.t.soka.ac.jp/java/DrawRings.html
LinkOut - more resources
Full Text Sources

