Automatic reconstruction of a bacterial regulatory network using Natural Language Processing
- PMID: 17683642
- PMCID: PMC1964768
- DOI: 10.1186/1471-2105-8-293
Automatic reconstruction of a bacterial regulatory network using Natural Language Processing
Abstract
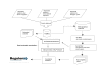
Background: Manual curation of biological databases, an expensive and labor-intensive process, is essential for high quality integrated data. In this paper we report the implementation of a state-of-the-art Natural Language Processing system that creates computer-readable networks of regulatory interactions directly from different collections of abstracts and full-text papers. Our major aim is to understand how automatic annotation using Text-Mining techniques can complement manual curation of biological databases. We implemented a rule-based system to generate networks from different sets of documents dealing with regulation in Escherichia coli K-12.
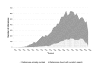
Results: Performance evaluation is based on the most comprehensive transcriptional regulation database for any organism, the manually-curated RegulonDB, 45% of which we were able to recreate automatically. From our automated analysis we were also able to find some new interactions from papers not already curated, or that were missed in the manual filtering and review of the literature. We also put forward a novel Regulatory Interaction Markup Language better suited than SBML for simultaneously representing data of interest for biologists and text miners.
Conclusion: Manual curation of the output of automatic processing of text is a good way to complement a more detailed review of the literature, either for validating the results of what has been already annotated, or for discovering facts and information that might have been overlooked at the triage or curation stages.
Figures

Similar articles
-
Extraction of biological interaction networks from scientific literature.Brief Bioinform. 2005 Sep;6(3):263-76. doi: 10.1093/bib/6.3.263. Brief Bioinform. 2005. PMID: 16212774 Review.
-
The comprehensive updated regulatory network of Escherichia coli K-12.BMC Bioinformatics. 2006 Jan 6;7:5. doi: 10.1186/1471-2105-7-5. BMC Bioinformatics. 2006. PMID: 16398937 Free PMC article.
-
Assisted curation of regulatory interactions and growth conditions of OxyR in E. coli K-12.Database (Oxford). 2014 Jun 4;2014:bau049. doi: 10.1093/database/bau049. Print 2014. Database (Oxford). 2014. PMID: 24903516 Free PMC article.
-
Wnt pathway curation using automated natural language processing: combining statistical methods with partial and full parse for knowledge extraction.Bioinformatics. 2005 Apr 15;21(8):1653-8. doi: 10.1093/bioinformatics/bti165. Epub 2004 Nov 25. Bioinformatics. 2005. PMID: 15564295
-
Hairpins in bookstacks: information retrieval from biomedical text.Brief Bioinform. 2005 Sep;6(3):222-38. doi: 10.1093/bib/6.3.222. Brief Bioinform. 2005. PMID: 16212771 Review.
Cited by
-
Reconstructing prokaryotic transcriptional regulatory networks: lessons from actinobacteria.J Biol. 2009;8(3):29. doi: 10.1186/jbiol132. Epub 2009 Apr 15. J Biol. 2009. PMID: 19435474 Free PMC article. Review.
-
Text mining and network analysis of molecular interaction in non-small cell lung cancer by using natural language processing.Mol Biol Rep. 2014 Dec;41(12):8071-9. doi: 10.1007/s11033-014-3705-5. Epub 2014 Sep 10. Mol Biol Rep. 2014. Retraction in: Mol Biol Rep. 2015 Oct;42(10):1489. doi: 10.1007/s11033-015-3908-4. PMID: 25205120 Retracted.
-
A text-mining system for extracting metabolic reactions from full-text articles.BMC Bioinformatics. 2012 Jul 23;13:172. doi: 10.1186/1471-2105-13-172. BMC Bioinformatics. 2012. PMID: 22823282 Free PMC article.
-
Overview of the gene regulation network and the bacteria biotope tasks in BioNLP'13 shared task.BMC Bioinformatics. 2015;16 Suppl 10(Suppl 10):S1. doi: 10.1186/1471-2105-16-S10-S1. Epub 2015 Jul 13. BMC Bioinformatics. 2015. PMID: 26202448 Free PMC article.
-
Linking genes to literature: text mining, information extraction, and retrieval applications for biology.Genome Biol. 2008;9 Suppl 2(Suppl 2):S8. doi: 10.1186/gb-2008-9-s2-s8. Epub 2008 Sep 1. Genome Biol. 2008. PMID: 18834499 Free PMC article. Review.
References
-
- Yandell MD, Majoros WH. Genomics and natural language processing. Nature Reviews – Genetics. 2002;3:601–10. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

