QTL mapping of domestication-related traits in soybean (Glycine max)
- PMID: 17684023
- PMCID: PMC2759197
- DOI: 10.1093/aob/mcm149
QTL mapping of domestication-related traits in soybean (Glycine max)
Abstract
Background and aims: Understanding the genetic basis underlying domestication-related traits (DRTs) is important in order to use wild germplasm efficiently for improving yield, stress tolerance and quality of crops. This study was conducted to characterize the genetic basis of DRTs in soybean (Glycine max) using quantitative trait locus (QTL) mapping.
Methods: A population of 96 recombinant inbred lines derived from a cultivated (ssp. max) x wild (ssp. soja) cross was used for mapping and QTL analysis. Nine DRTs were examined in 2004 and 2005. A linkage map was constructed with 282 markers by the Kosambi function, and the QTL was detected by composite interval mapping.
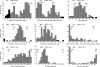
Key results: The early flowering and determinate habit derived from the max parent were each controlled by one major QTL, corresponding to the major genes for maturity (e1) and determinate habit (dt1), respectively. There were only one or two significant QTLs for twinning habit, pod dehiscence, seed weight and hard seededness, which each accounted for approx. 20-50 % of the total variance. A comparison with the QTLs detected previously indicated that in pod dehiscence and hard seededness, at least one major QTL was common across different crosses, whereas no such consistent QTL existed for seed weight.
Conclusions: Most of the DRTs in soybeans were conditioned by one or two major QTLs and a number of genotype-dependent minor QTLs. The common major QTLs identified in pod dehiscence and hard seededness may have been key loci in the domestication of soybean. The evolutionary changes toward larger seed may have occurred through the accumulation of minor changes at many QTLs. Since the major QTLs for DRTs were scattered across only six of the 20 linkage groups, and since the QTLs were not clustered, introgression of useful genes from wild to cultivated soybeans can be carried out without large obstacles.
Figures
Similar articles
-
Genetic dissection of domestication-related traits in soybean through genotyping-by-sequencing of two interspecific mapping populations.Theor Appl Genet. 2019 Apr;132(4):1195-1209. doi: 10.1007/s00122-018-3272-6. Epub 2019 Jan 3. Theor Appl Genet. 2019. PMID: 30607438
-
QTL mapping pod dehiscence resistance in soybean (Glycine max L. Merr.) using specific-locus amplified fragment sequencing.Theor Appl Genet. 2019 Aug;132(8):2253-2272. doi: 10.1007/s00122-019-03352-x. Epub 2019 Jun 3. Theor Appl Genet. 2019. PMID: 31161230 Free PMC article.
-
Construction of a high-density genetic map and mapping of QTLs for soybean (Glycine max) agronomic and seed quality traits by specific length amplified fragment sequencing.BMC Genomics. 2018 Aug 29;19(1):641. doi: 10.1186/s12864-018-5035-9. BMC Genomics. 2018. PMID: 30157757 Free PMC article.
-
Divergence of flowering genes in soybean.J Biosci. 2012 Nov;37(5):857-70. doi: 10.1007/s12038-012-9252-0. J Biosci. 2012. PMID: 23107921 Review.
-
[Occurrence characteristics and molecular genetic basis of pod shattering in soybean].Yi Chuan. 2015 Jun;37(6):535-43. doi: 10.16288/j.yczz.14-456. Yi Chuan. 2015. PMID: 26351049 Review. Chinese.
Cited by
-
Genetic and molecular bases of photoperiod responses of flowering in soybean.Breed Sci. 2012 Jan;61(5):531-43. doi: 10.1270/jsbbs.61.531. Epub 2012 Feb 4. Breed Sci. 2012. PMID: 23136492 Free PMC article.
-
Identification of major quantitative trait loci and candidate genes for seed weight in soybean.Theor Appl Genet. 2023 Jan;136(1):22. doi: 10.1007/s00122-023-04299-w. Epub 2023 Jan 23. Theor Appl Genet. 2023. PMID: 36688967 Free PMC article.
-
Quantitative genetic analysis of embryo heterosis in faba bean (Vicia faba L.).Theor Appl Genet. 2010 Jan;120(2):261-70. doi: 10.1007/s00122-009-1057-7. Theor Appl Genet. 2010. PMID: 19449175 Free PMC article.
-
Genome-wide identification and evolutionary analysis of leucine-rich repeat receptor-like protein kinase genes in soybean.BMC Plant Biol. 2016 Mar 2;16:58. doi: 10.1186/s12870-016-0744-1. BMC Plant Biol. 2016. PMID: 26935840 Free PMC article.
-
Unravelling the genetic architecture of soybean tofu quality traits.Mol Breed. 2025 Jan 3;45(1):8. doi: 10.1007/s11032-024-01529-x. eCollection 2025 Jan. Mol Breed. 2025. PMID: 39758754 Free PMC article.
References
-
- Abe J, Ohara M, Shimamoto Y. New electrophoretic mobility variants observed in wild soybean (Glycine soja) distributed in Japan and Korea. Soybean Genetics Newsletter. 1992;19:63–72.
-
- Abe J, Hasegawa A, Fukushi H, Mikami T, Ohara M, Shimamoto Y. Introgression between wild and cultivated soybeans of Japan revealed by RFLP analysis of chloroplast DNAs. Economic Botany. 1999;53:285–291.
-
- Bailey MA, Mian MAR, Carter TE, Jr, Ashley DA, Boerma HR. Pod dehiscence of soybean: identification of quantitative trait loci. Journal of Heredity. 1997;88:152–154.
-
- Box GEP, Cox DR. An analysis of transformations. Journal of the Royal Statistics Society, Series B. 1964;26:211–243.
-
- Broich S, Palmer RG. A cluster analysis of wild and domesticated soybean phenotypes. Euphytica. 1980;29:23–32.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

