Extensive gains and losses of olfactory receptor genes in mammalian evolution
- PMID: 17684554
- PMCID: PMC1933591
- DOI: 10.1371/journal.pone.0000708
Extensive gains and losses of olfactory receptor genes in mammalian evolution
Abstract
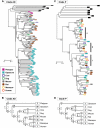
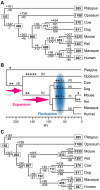
Odor perception in mammals is mediated by a large multigene family of olfactory receptor (OR) genes. The number of OR genes varies extensively among different species of mammals, and most species have a substantial number of pseudogenes. To gain some insight into the evolutionary dynamics of mammalian OR genes, we identified the entire set of OR genes in platypuses, opossums, cows, dogs, rats, and macaques and studied the evolutionary change of the genes together with those of humans and mice. We found that platypuses and primates have <400 functional OR genes while the other species have 800-1,200 functional OR genes. We then estimated the numbers of gains and losses of OR genes for each branch of the phylogenetic tree of mammals. This analysis showed that (i) gene expansion occurred in the placental lineage each time after it diverged from monotremes and from marsupials and (ii) hundreds of gains and losses of OR genes have occurred in an order-specific manner, making the gene repertoires highly variable among different orders. It appears that the number of OR genes is determined primarily by the functional requirement for each species, but once the number reaches the required level, it fluctuates by random duplication and deletion of genes. This fluctuation seems to have been aided by the stochastic nature of OR gene expression.
Conflict of interest statement
Figures
References
-
- Buck L, Axel R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell. 1991;65:175–187. - PubMed
-
- Mombaerts P. Genes and ligands for odorant, vomeronasal and taste receptors. Nat Rev Neurosci. 2004;5:263–278. - PubMed
-
- Glusman G, Yanai I, Rubin I, Lancet D. The complete human olfactory subgenome. Genome Res. 2001;11:685–702. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

