Quantitative sequence-function relationships in proteins based on gene ontology
- PMID: 17686158
- PMCID: PMC1976327
- DOI: 10.1186/1471-2105-8-294
Quantitative sequence-function relationships in proteins based on gene ontology
Abstract
Background: The relationship between divergence of amino-acid sequence and divergence of function among homologous proteins is complex. The assumption that homologs share function--the basis of transfer of annotations in databases--must therefore be regarded with caution. Here, we present a quantitative study of sequence and function divergence, based on the Gene Ontology classification of function. We determined the relationship between sequence divergence and function divergence in 6828 protein families from the PFAM database. Within families there is a broad range of sequence similarity from very closely related proteins--for instance, orthologs in different mammals--to very distantly-related proteins at the limit of reliable recognition of homology.
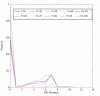
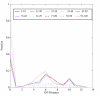
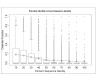
Results: We correlated the divergence in sequences determined from pairwise alignments, and the divergence in function determined by path lengths in the Gene Ontology graph, taking into account the fact that many proteins have multiple functions. Our results show that, among homologous proteins, the proportion of divergent functions decreases dramatically above a threshold of sequence similarity at about 50% residue identity. For proteins with more than 50% residue identity, transfer of annotation between homologs will lead to an erroneous attribution with a totally dissimilar function in fewer than 6% of cases. This means that for very similar proteins (about 50 % identical residues) the chance of completely incorrect annotation is low; however, because of the phenomenon of recruitment, it is still non-zero.
Conclusion: Our results describe general features of the evolution of protein function, and serve as a guide to the reliability of annotation transfer, based on the closeness of the relationship between a new protein and its nearest annotated relative.
Figures







Similar articles
-
SUPFAM--a database of potential protein superfamily relationships derived by comparing sequence-based and structure-based families: implications for structural genomics and function annotation in genomes.Nucleic Acids Res. 2002 Jan 1;30(1):289-93. doi: 10.1093/nar/30.1.289. Nucleic Acids Res. 2002. PMID: 11752317 Free PMC article.
-
On the quality of tree-based protein classification.Bioinformatics. 2005 May 1;21(9):1876-90. doi: 10.1093/bioinformatics/bti244. Epub 2005 Jan 12. Bioinformatics. 2005. PMID: 15647305
-
Evolutionary rates at codon sites may be used to align sequences and infer protein domain function.BMC Bioinformatics. 2010 Mar 24;11:151. doi: 10.1186/1471-2105-11-151. BMC Bioinformatics. 2010. PMID: 20334658 Free PMC article.
-
Protein Function Prediction: Problems and Pitfalls.Curr Protoc Bioinformatics. 2015 Sep 3;51:4.12.1-4.12.8. doi: 10.1002/0471250953.bi0412s51. Curr Protoc Bioinformatics. 2015. PMID: 26334923 Review.
-
Pairwise statistical significance and empirical determination of effective gap opening penalties for protein local sequence alignment.Int J Comput Biol Drug Des. 2008;1(4):347-67. doi: 10.1504/ijcbdd.2008.022207. Int J Comput Biol Drug Des. 2008. PMID: 20063463 Review.
Cited by
-
Resolving the ortholog conjecture: orthologs tend to be weakly, but significantly, more similar in function than paralogs.PLoS Comput Biol. 2012;8(5):e1002514. doi: 10.1371/journal.pcbi.1002514. Epub 2012 May 17. PLoS Comput Biol. 2012. PMID: 22615551 Free PMC article.
-
SPOT: A machine learning model that predicts specific substrates for transport proteins.PLoS Biol. 2024 Sep 26;22(9):e3002807. doi: 10.1371/journal.pbio.3002807. eCollection 2024 Sep. PLoS Biol. 2024. PMID: 39325691 Free PMC article.
-
Stromal microenvironment processes unveiled by biological component analysis of gene expression in xenograft tumor models.BMC Bioinformatics. 2010 Oct 28;11 Suppl 9(Suppl 9):S11. doi: 10.1186/1471-2105-11-S9-S11. BMC Bioinformatics. 2010. PMID: 21044358 Free PMC article.
-
Searching the protein structure database for ligand-binding site similarities using CPASS v.2.BMC Res Notes. 2011 Jan 26;4:17. doi: 10.1186/1756-0500-4-17. BMC Res Notes. 2011. PMID: 21269480 Free PMC article.
-
Evolutionary innovations and the organization of protein functions in genotype space.PLoS One. 2010 Nov 30;5(11):e14172. doi: 10.1371/journal.pone.0014172. PLoS One. 2010. PMID: 21152394 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

