Chromosome numbers and genome size variation in Indian species of Curcuma (Zingiberaceae)
- PMID: 17686760
- PMCID: PMC2533610
- DOI: 10.1093/aob/mcm144
Chromosome numbers and genome size variation in Indian species of Curcuma (Zingiberaceae)
Abstract
Background and aims: Genome size and chromosome numbers are important cytological characters that significantly influence various organismal traits. However, geographical representation of these data is seriously unbalanced, with tropical and subtropical regions being largely neglected. In the present study, an investigation was made of chromosomal and genome size variation in the majority of Curcuma species from the Indian subcontinent, and an assessment was made of the value of these data for taxonomic purposes.
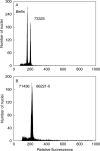
Methods: Genome size of 161 homogeneously cultivated plant samples classified into 51 taxonomic entities was determined by propidium iodide flow cytometry. Chromosome numbers were counted in actively growing root tips using conventional rapid squash techniques.
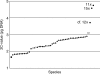
Key results: Six different chromosome counts (2n = 22, 42, 63, >70, 77 and 105) were found, the last two representing new generic records. The 2C-values varied from 1.66 pg in C. vamana to 4.76 pg in C. oligantha, representing a 2.87-fold range. Three groups of taxa with significantly different homoploid genome sizes (Cx-values) and distinct geographical distribution were identified. Five species exhibited intraspecific variation in nuclear DNA content, reaching up to 15.1 % in cultivated C. longa. Chromosome counts and genome sizes of three Curcuma-like species (Hitchenia caulina, Kaempferia scaposa and Paracautleya bhatii) corresponded well with typical hexaploid (2n = 6x = 42) Curcuma spp.
Conclusions: The basic chromosome number in the majority of Indian taxa (belonging to subgenus Curcuma) is x = 7; published counts correspond to 6x, 9x, 11x, 12x and 15x ploidy levels. Only a few species-specific C-values were found, but karyological and/or flow cytometric data may support taxonomic decisions in some species alliances with morphological similarities. Close evolutionary relationships among some cytotypes are suggested based on the similarity in homoploid genome sizes and geographical grouping. A new species combination, Curcuma scaposa (Nimmo) Skornick. & M. Sabu, comb. nov., is proposed.
Figures

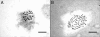

References
-
- Apavatjrut P, Sirisawad T, Sirirugsa P, Voraurai P, Suwanthada C. Studies on chromosome number of seventeen Thai Curcuma species. Proceedings of 2nd National Conference on Flower and Ornamental Plant; 1996. pp. 86–99.
-
- Ardiyani M. Systematic study of Curcuma L.: turmeric and its allies. Scotland: University of Edinburgh; 2002. PhD thesis.
-
- Beltran IC, Kam YK. Cytotaxonomic studies in the Zingiberaceae. Notes from the Royal Botanic Garden Edinburgh. 1984;41:541–559.
-
- Bennett MD, Leitch IJ. Plant DNA C-values Database (Release 4·0, Oct. 2005) 2005a. http://www.kew.org/genomesize/homepage. (accessed 5 January, 2007).
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

