A survey of bacterial insertion sequences using IScan
- PMID: 17686783
- PMCID: PMC2018620
- DOI: 10.1093/nar/gkm597
A survey of bacterial insertion sequences using IScan
Abstract
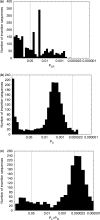
Bacterial insertion sequences (ISs) are the simplest kinds of bacterial mobile DNA. Evolutionary studies need consistent IS annotation across many different genomes. We have developed an open-source software package, IScan, to identify bacterial ISs and their sequence elements--inverted and target direct repeats--in multiple genomes using multiple flexible search parameters. We applied IScan to 438 completely sequenced bacterial genomes and 20 IS families. The resulting data show that ISs within a genome are extremely similar, with a mean synonymous divergence of K(s) = 0.033. Our analysis substantially extends previously available information, and suggests that most ISs have entered bacterial genomes recently. By implication, their population persistence may depend on horizontal transfer. We also used IScan's ability to analyze the statistical significance of sequence similarity among many IS inverted repeats. Although the inverted repeats of insertion sequences are evolutionarily highly flexible parts of ISs, we show that this ability can be used to enrich a dataset for ISs that are likely to be functional. Applied to the thousands of genomes that will soon be available, IScan could be used for many purposes, such as mapping the evolutionary history and horizontal transfer patterns of different ISs.
Figures
References
-
- Siguier P, Filee J, Chandler M. Insertion sequences in prokaryotic genomes. Curr. Opin. Microbiol. 2006;9:526–531. - PubMed
-
- Lohe AR, Moriyama EN, Lidholm DA, Hartl DL. Horizontal transmission, vertical inactivation, and stochastic loss of mariner-like transposable elements. Mol. Biol. Evol. 1995;12:62–72. - PubMed
-
- Capy P, Langin T, Bigot Y, Brunet F, Daboussi MJ, Periquet G, David JR, Hartl DL. Horizontal transmission versus ancient origin - mariner in the witness box. Genetica. 1994;93:161–170. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

