DNA palindromes with a modest arm length of greater, similar 20 base pairs are a significant target for recombinant adeno-associated virus vector integration in the liver, muscles, and heart in mice
- PMID: 17686840
- PMCID: PMC2045527
- DOI: 10.1128/JVI.00963-07
DNA palindromes with a modest arm length of greater, similar 20 base pairs are a significant target for recombinant adeno-associated virus vector integration in the liver, muscles, and heart in mice
Abstract
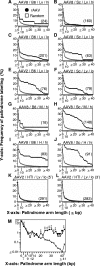
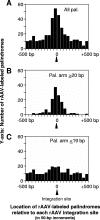
Our previous study has shown that recombinant adeno-associated virus (rAAV) vector integrates preferentially in genes, near transcription start sites and CpG islands in mouse liver (H. Nakai, X. Wu, S. Fuess, T. A. Storm, D. Munroe, E. Montini, S. M. Burgess, M. Grompe, and M. A. Kay, J. Virol. 79:3606-3614, 2005). However, the previous method relied on in vivo selection of rAAV integrants and could be employed for the liver but not for other tissues. Here, we describe a novel method for high-throughput rAAV integration site analysis that does not rely on marker gene expression, selection, or cell division, and therefore it can identify rAAV integration sites in nondividing cells without cell manipulations. Using this new method, we identified and characterized a total of 997 rAAV integration sites in mouse liver, skeletal muscle, and heart, transduced with rAAV2 or rAAV8 vector. The results support our previous observations, but notably they have revealed that DNA palindromes with an arm length of greater, similar 20 bp (total length, greater, similar 40 bp) are a significant target for rAAV integration. Up to approximately 30% of total integration events occurred in the vicinity of DNA palindromes with an arm length of greater, similar 20 bp. Considering that DNA palindromes may constitute fragile genomic sites, our results support the notion that rAAV integrates at chromosomal sites susceptible to breakage or preexisting breakage sites. The use of rAAV to label fragile genomic sites may provide an important new tool for probing the intrinsic source of ongoing genomic instability in various tissues in animals, studying DNA palindrome metabolism in vivo, and understanding their possible contributions to carcinogenesis and aging.
Figures



References
-
- Butler, D. K., L. E. Yasuda, and M. C. Yao. 1996. Induction of large DNA palindrome formation in yeast: implications for gene amplification and genome stability in eukaryotes. Cell 87:1115-1122. - PubMed
-
- Donsante, A., C. Vogler, N. Muzyczka, J. M. Crawford, J. Barker, T. Flotte, M. Campbell-Thompson, T. Daly, and M. S. Sands. 2001. Observed incidence of tumorigenesis in long-term rodent studies of rAAV vectors. Gene Ther. 8:1343-1346. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

