Lethal mutagenesis of poliovirus mediated by a mutagenic pyrimidine analogue
- PMID: 17686844
- PMCID: PMC2045539
- DOI: 10.1128/JVI.01028-07
Lethal mutagenesis of poliovirus mediated by a mutagenic pyrimidine analogue
Abstract
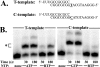
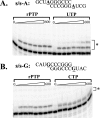
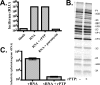
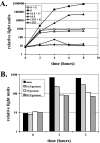
Lethal mutagenesis is the mechanism of action of ribavirin against poliovirus (PV) and numerous other RNA viruses. However, there is still considerable debate regarding the mechanism of action of ribavirin against a variety of RNA viruses. Here we show by using T7 RNA polymerase-mediated production of PV genomic RNA, PV polymerase-catalyzed primer extension, and cell-free PV synthesis that a pyrimidine ribonucleoside triphosphate analogue (rPTP) with ambiguous base-pairing capacity is an efficient mutagen of the PV genome. The in vitro incorporation properties of rPTP are superior to ribavirin triphosphate. We observed a log-linear relationship between virus titer reduction and the number of rPMP molecules incorporated. A PV genome encoding a high-fidelity polymerase was more sensitive to rPMP incorporation, consistent with diminished mutational robustness of high-fidelity PV. The nucleoside (rP) did not exhibit antiviral activity in cell culture, owing to the inability of rP to be converted to rPMP by cellular nucleotide kinases. rP was also a poor substrate for herpes simplex virus thymidine kinase. The block to nucleoside phosphorylation could be bypassed by treatment with the P nucleobase, which exhibited both antiviral activity and mutagenesis, presumably a reflection of rP nucleotide formation by a nucleotide salvage pathway. These studies provide additional support for lethal mutagenesis as an antiviral strategy, suggest that rPMP prodrugs may be highly efficacious antiviral agents, and provide a new tool to determine the sensitivity of RNA virus genomes to mutagenesis as well as interrogation of the impact of mutational load on the population dynamics of these viruses.
Figures
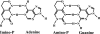
Similar articles
-
Lethal mutagenesis of picornaviruses with N-6-modified purine nucleoside analogues.Antimicrob Agents Chemother. 2008 Mar;52(3):971-9. doi: 10.1128/AAC.01056-07. Epub 2008 Jan 7. Antimicrob Agents Chemother. 2008. PMID: 18180344 Free PMC article.
-
Ribavirin and lethal mutagenesis of poliovirus: molecular mechanisms, resistance and biological implications.Virus Res. 2005 Feb;107(2):173-81. doi: 10.1016/j.virusres.2004.11.007. Virus Res. 2005. PMID: 15649563 Review.
-
The broad-spectrum antiviral ribonucleoside ribavirin is an RNA virus mutagen.Nat Med. 2000 Dec;6(12):1375-9. doi: 10.1038/82191. Nat Med. 2000. PMID: 11100123
-
A single mutation in poliovirus RNA-dependent RNA polymerase confers resistance to mutagenic nucleotide analogs via increased fidelity.Proc Natl Acad Sci U S A. 2003 Jun 10;100(12):7289-94. doi: 10.1073/pnas.1232294100. Epub 2003 May 16. Proc Natl Acad Sci U S A. 2003. PMID: 12754380 Free PMC article.
-
Lethal Mutagenesis of RNA Viruses and Approved Drugs with Antiviral Mutagenic Activity.Viruses. 2022 Apr 18;14(4):841. doi: 10.3390/v14040841. Viruses. 2022. PMID: 35458571 Free PMC article. Review.
Cited by
-
Anti-HIV-1 activity of resveratrol derivatives and synergistic inhibition of HIV-1 by the combination of resveratrol and decitabine.Bioorg Med Chem Lett. 2012 Nov 1;22(21):6642-6. doi: 10.1016/j.bmcl.2012.08.108. Epub 2012 Sep 6. Bioorg Med Chem Lett. 2012. PMID: 23010273 Free PMC article.
-
Fidelity variants of RNA dependent RNA polymerases uncover an indirect, mutagenic activity of amiloride compounds.PLoS Pathog. 2010 Oct 28;6(10):e1001163. doi: 10.1371/journal.ppat.1001163. PLoS Pathog. 2010. PMID: 21060812 Free PMC article.
-
Lethal mutagenesis of bacteria.Genetics. 2008 Oct;180(2):1061-70. doi: 10.1534/genetics.108.091413. Epub 2008 Sep 9. Genetics. 2008. PMID: 18780744 Free PMC article.
-
Does mutational robustness inhibit extinction by lethal mutagenesis in viral populations?PLoS Comput Biol. 2010 Jun 10;6(6):e1000811. doi: 10.1371/journal.pcbi.1000811. PLoS Comput Biol. 2010. PMID: 20548958 Free PMC article.
-
Quasispecies theory for horizontal gene transfer and recombination.Phys Rev E Stat Nonlin Soft Matter Phys. 2008 Dec;78(6 Pt 1):061921. doi: 10.1103/PhysRevE.78.061921. Epub 2008 Dec 23. Phys Rev E Stat Nonlin Soft Matter Phys. 2008. PMID: 19256882 Free PMC article.
References
-
- Airaksinen, A., N. Pariente, L. Menendez-Arias, and E. Domingo. 2003. Curing of foot-and-mouth disease virus from persistently infected cells by ribavirin involves enhanced mutagenesis. Virology 311:339-349. - PubMed
-
- Arnold, J. J., and C. E. Cameron. 2000. Poliovirus RNA-dependent RNA polymerase (3Dpol). Assembly of stable, elongation-competent complexes by using a symmetrical primer-template substrate (sym/sub). J. Biol. Chem. 275:5329-5336. - PubMed
-
- Crain, P. F. 1990. Preparation and enzymatic hydrolysis of DNA and RNA for mass spectrometry. Methods Enzymol. 193:782-790. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

