Reconstitution of a human ATR-mediated checkpoint response to damaged DNA
- PMID: 17686975
- PMCID: PMC1941640
- DOI: 10.1073/pnas.0706013104
Reconstitution of a human ATR-mediated checkpoint response to damaged DNA
Abstract
The DNA damage checkpoint response delays cell cycle progression upon DNA damage and prevents genomic instability. Genetic analysis has identified sensor, mediator, signal transducer, and effector components of this global signal transduction pathway. Here we describe an in vitro system with purified human checkpoint proteins that recapitulates key elements of the DNA damage checkpoint. We show that the damage sensor ATR in the presence of topoisomerase II binding protein 1 (TopBP1) mediator/adaptor protein phosphorylates the Chk1 signal-transducing kinase in a reaction that is strongly dependent on the presence of DNA containing bulky base lesions. The dependence on damaged DNA requires DNA binding by TopBP1, and, indeed, TopBP1 shows preferential binding to damaged DNA. This in vitro system provides a useful platform for mechanistic studies of the human DNA damage checkpoint response.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

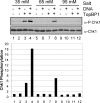
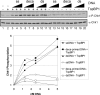

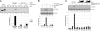
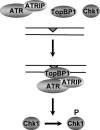
References
-
- Abraham RT. Genes Dev. 2001;15:2177–2196. - PubMed
-
- Nyberg KA, Michelson RJ, Putnam CW, Weinert TA. Annu Rev Genet. 2002;36:617–656. - PubMed
-
- Sancar A, Lindsey-Boltz LA, Ünsal-Kaçmaz K, Linn S. Annu Rev Biochem. 2004;73:39–85. - PubMed
-
- Lee JH, Paull TT. Science. 2004;304:93–96. - PubMed
-
- Lee JH, Paull TT. Science. 2005;308:551–554. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

