Structural basis for the binding of the neutralizing antibody, 7D11, to the poxvirus L1 protein
- PMID: 17688903
- PMCID: PMC2100026
- DOI: 10.1016/j.virol.2007.06.042
Structural basis for the binding of the neutralizing antibody, 7D11, to the poxvirus L1 protein
Abstract
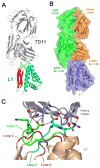
Medical countermeasures to prevent or treat smallpox are needed due to the potential use of poxviruses as biological weapons. Safety concerns with the currently available smallpox vaccine indicate a need for research on alternative poxvirus vaccine strategies. Molecular vaccines involving the use of proteins and/or genes and recombinant antibodies are among the strategies under current investigation. The poxvirus L1 protein, encoded by the L1R open reading frame, is the target of neutralizing antibodies and has been successfully used as a component of both protein subunit and DNA vaccines. L1-specific monoclonal antibodies (e.g., mouse monoclonal antibody mAb-7D11, mAb-10F5) with potent neutralizing activity bind L1 in a conformation-specific manner. This suggests that proper folding of the L1 protein used in molecular vaccines will affect the production of neutralizing antibodies and protection. Here, we co-crystallized the Fab fragment of mAb-7D11 with the L1 protein. The crystal structure of the complex between Fab-7D11 and L1 reveals the basis for the conformation-specific binding as recognition of a discontinuous epitope containing two loops that are held together by a disulfide bond. The structure of this important conformational epitope of L1 will contribute to the development of molecular poxvirus vaccines and also provides a novel target for anti-poxvirus drugs. In addition, the sequence and structure of Fab-7D11 will contribute to the development of L1-targeted immunotherapeutics.
Figures


References
-
- Aldaz-Carroll L, Whitbeck J, Ponce De Leon M, Lou H, Pannell L, Lebowitz J, Fogg C, White CL, Moss B, Cohen GH, Eisenberg R. Physical and immunological characterization of a recombinant secreted form of the membrane protein encoded by the vaccinia virus L1R gene. Virology. 2005;341(1):59–71. - PubMed
-
- Chen Z, Earl P, Americo J, Damon I, Smith SK, Zhou Y, Yu F, Sebrell A, Emerson S, Cohen G, Eisenberg R, Svitel J, Schuck P, Satterfield W, Moss B, Purcell R. Chimpanzee/human mAbs to vaccinia virus B5 protein neutralize vaccinia and smallpox viruses and protect mice against vaccinia virus. Proc Natl Acad Sci USA. 2006;103(6):1882–1887. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

