Cloning and functional characterization of a novel mitochondrial N-ethylmaleimide-sensitive glycerol-3-phosphate acyltransferase (GPAT2)
- PMID: 17689486
- PMCID: PMC2133398
- DOI: 10.1016/j.abb.2007.06.033
Cloning and functional characterization of a novel mitochondrial N-ethylmaleimide-sensitive glycerol-3-phosphate acyltransferase (GPAT2)
Abstract
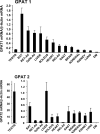
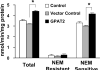
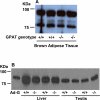
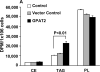
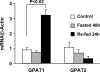
Glycerol-3-phosphate acyltransferase (GPAT) catalyzes the initial and rate-limiting step in glycerolipid synthesis. Several mammalian GPAT activities have been recognized, including N-ethylmaleimide (NEM)-sensitive isoforms in microsomes and mitochondria and an NEM-resistant form in mitochondrial outer membrane (GPAT1). We have now cloned a second mitochondrial isoform, GPAT2 from mouse testis. The open-reading frame encodes a protein of 798 amino acids with a calculated mass of 88.8kDa and 27% amino acid identity to GPAT1. Testis mRNA expression was 50-fold higher than in liver or brown adipose tissue, but the specific activity of NEM-sensitive GPAT in testis mitochondria was similar to that in liver. When Cos-7 cells were transiently transfected with GPAT2, NEM-sensitive GPAT activity increased 30%. Confocal microscopy confirmed a mitochondrial location. Incubation of GPAT2-transfected Cos-7 cells with trace (3 microM; 0.25 microCi) [1-(14)C]oleate for 6h increased incorporation of [(14)C]oleate into TAG 84%. In contrast, incorporation into phospholipid species was lower than in control cells. Although a polyclonal antibody raised against full-length GPAT1 detected an approximately 89-kDa band in liver and testis from GPAT1 null mice and both 89- and 80-kDa bands in BAT from the knockout animals, the GPAT2 protein expressed in Cos-7 cells was only 80 kDa. In vitro translation showed a single product of 89 kDa. Unlike GPAT1, GPAT2 mRNA abundance in liver was not altered by fasting or refeeding. GPAT2 is likely to have a specialized function in testis.
Figures







References
-
- Coleman RA, Lee DP. Enzymes of triacylglycerol synthesis and their regulation. Prog. Lipid Res. 2004;43:134–176. - PubMed
-
- Lewin TM, Schwerbrock NMJ, Lee DP, Coleman RA. Identification of a new glycerol-3-phosphate acyltransferase isoenzyme, mtGPAT2, in mitochondria. J. Biol. Chem. 2004;279:13488–13495. - PubMed
-
- Vancura A, Haldar D. Purification and characterization of glycerophosphate acyltransferase from rat liver mitochondria. J. Biol. Chem. 1994;269:27209–27215. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

