Phospho-epitope binding by the BRCT domains of hPTIP controls multiple aspects of the cellular response to DNA damage
- PMID: 17690115
- PMCID: PMC2018624
- DOI: 10.1093/nar/gkm493
Phospho-epitope binding by the BRCT domains of hPTIP controls multiple aspects of the cellular response to DNA damage
Erratum in
-
Corrigendum: Phospho-epitope binding by the BRCT domains of hPTIP controls multiple aspects of the cellular response to DNA damage.Nucleic Acids Res. 2017 Dec 15;45(22):13094. doi: 10.1093/nar/gkx1140. Nucleic Acids Res. 2017. PMID: 29136188 Free PMC article. No abstract available.
Expression of concern in
-
Editor's note to 'phospho-epitope binding by the BRCT domains of hPTIP controls multiple aspects of the cellular response to DNA damage'.Nucleic Acids Res. 2022 Nov 28;50(21):12599. doi: 10.1093/nar/gkac1165. Nucleic Acids Res. 2022. PMID: 36440756 Free PMC article. No abstract available.
Abstract
Human (h)PTIP plays important but poorly understood roles in cellular responses to DNA damage. hPTIP interacts with 53BP1 tumour suppressor but only when 53BP1 is phosphorylated by ATM after DNA damage although the mechanism(s) and significance of the interaction of these two proteins are unclear. Here, we pinpoint a single ATM-phosphorylated residue in 53BP1--Ser25--that is required for binding of 53BP1 to hPTIP. Binding of phospho-Ser25 to hPTIP in vitro and in vivo requires two closely apposed pairs of BRCT domains at the C-terminus of hPTIP and neither pair alone can bind to phospho-Ser25, even though one of these BRCT pairs in isolation can bind to other ATM-phosphorylated epitopes. Mutations in 53BP1 and in hPTIP that prevent the interaction of the two proteins, render cells hypersensitive to DNA damage and weaken ATM signalling. The C-terminal BRCT domains of hPTIP are also required for stable retention of hPTIP at sites of DNA damage but this appears to be independent of binding to 53BP1. Thus, the BRCT domains of hPTIP play important roles in the cellular response to DNA damage.
Figures
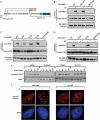
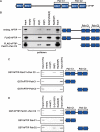
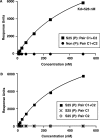
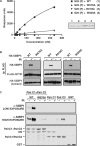

References
-
- Jowsey PA, Doherty AJ, Rouse J. Human PTIP facilitates ATM-mediated activation of p53 and promotes cellular resistance to ionizing radiation. J. Biol. Chem. 2004;279:55562–55569. - PubMed
-
- Manke IA, Lowery DM, Nguyen A, Yaffe MB. BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science. 2003;302:636–639. - PubMed
-
- Durocher D, Jackson SP. DNA-PK, ATM and ATR as sensors of DNA damage: variations on a theme? Curr. Opin. Cell. Biol. 2001;13:225–231. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

