Cdc18/CDC6 activates the Rad3-dependent checkpoint in the fission yeast
- PMID: 17690116
- PMCID: PMC2018612
- DOI: 10.1093/nar/gkm527
Cdc18/CDC6 activates the Rad3-dependent checkpoint in the fission yeast
Abstract
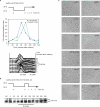
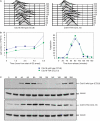
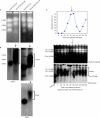
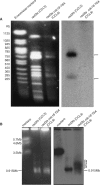
A screen for genes that can ectopically activate a Rad3-dependent checkpoint block over mitosis in fission yeast has identified the DNA replication initiation factor cdc18 (known as CDC6 in other organisms). Either a stabilized form of Cdc18, the Cdc18-T6A phosphorylation mutant, or overexpression of wild type Cdc18, activate the Rad3-dependent S-M checkpoint in the apparent absence of detectable replication structures and gross DNA damage. This cell cycle block relies on the Rad checkpoint pathway and requires Chk1 phosphorylation and activation. Unexpectedly, Cdc18-T6A induces changes in the mobility of Chromosome III, affecting the size of a restriction fragment containing rDNA repeats and producing aberrant nucleolar structures. Recombination events within the rDNA appear to contribute at least in part to the cell cycle delay. We propose that an elevated level of Cdc18 activates the Rad3-dependent checkpoint either directly or indirectly, and additionally causes expansion of the rDNA repeats on Chromosome III.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

