Whole proteome analysis of post-translational modifications: applications of mass-spectrometry for proteogenomic annotation
- PMID: 17690205
- PMCID: PMC1950905
- DOI: 10.1101/gr.6427907
Whole proteome analysis of post-translational modifications: applications of mass-spectrometry for proteogenomic annotation
Abstract
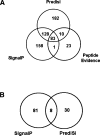
While bacterial genome annotations have significantly improved in recent years, techniques for bacterial proteome annotation (including post-translational chemical modifications, signal peptides, proteolytic events, etc.) are still in their infancy. At the same time, the number of sequenced bacterial genomes is rising sharply, far outpacing our ability to validate the predicted genes, let alone annotate bacterial proteomes. In this study, we use tandem mass spectrometry (MS/MS) to annotate the proteome of Shewanella oneidensis MR-1, an important microbe for bioremediation. In particular, we provide the first comprehensive map of post-translational modifications in a bacterial genome, including a large number of chemical modifications, signal peptide cleavages, and cleavages of N-terminal methionine residues. We also detect multiple genes that were missed or assigned incorrect start positions by gene prediction programs, and suggest corrections to improve the gene annotation. This study demonstrates that complementing every genome sequencing project by an MS/MS project would significantly improve both genome and proteome annotations for a reasonable cost.
Figures











References
-
- Aebersold R., Mann M., Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. - PubMed
-
- Antelmann H., Tjalsma H., Voight B., Ohlmeier S., Bron S., van Dijl J.M., Hecker M., Tjalsma H., Voight B., Ohlmeier S., Bron S., van Dijl J.M., Hecker M., Voight B., Ohlmeier S., Bron S., van Dijl J.M., Hecker M., Ohlmeier S., Bron S., van Dijl J.M., Hecker M., Bron S., van Dijl J.M., Hecker M., van Dijl J.M., Hecker M., Hecker M. A proteomic view on genome-based signal peptide predictions. Genome Res. 2001;11:1484–1502. - PubMed
-
- Arnold R.J., Reilly J.P., Reilly J.P. Observation of Escherichia coli ribosomal proteins and their posttranslational modifications by mass spectrometry. Anal. Biochem. 1999;269:105–112. - PubMed
-
- Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K., Tomita M., Wanner B., Mori H., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K., Tomita M., Wanner B., Mori H., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K., Tomita M., Wanner B., Mori H., Takai Y., Okumura Y., Baba M., Datsenko K., Tomita M., Wanner B., Mori H., Okumura Y., Baba M., Datsenko K., Tomita M., Wanner B., Mori H., Baba M., Datsenko K., Tomita M., Wanner B., Mori H., Datsenko K., Tomita M., Wanner B., Mori H., Tomita M., Wanner B., Mori H., Wanner B., Mori H., Mori H., et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol. Syst. Biol. 2006;2:E1–E11. doi: 10.1038/msb4100050. - DOI - PMC - PubMed
-
- Baranov P., Gesteland R., Atkins J., Gesteland R., Atkins J., Atkins J. Recoding: Translational bifurcations in gene expression. Gene. 2002;286:187–201. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
