The Ras-association domain family (RASSF) members and their role in human tumourigenesis
- PMID: 17692468
- PMCID: PMC2586335
- DOI: 10.1016/j.bbcan.2007.06.003
The Ras-association domain family (RASSF) members and their role in human tumourigenesis
Abstract
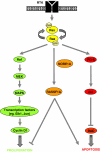
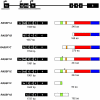
Ras proteins play a direct causal role in human cancer with activating mutations in Ras occurring in approximately 30% of tumours. Ras effectors also contribute to cancer, as mutations occur in Ras effectors, notably B-Raf and PI3-K, and drugs blocking elements of these pathways are in clinical development. In 2000, a new Ras effector was identified, RAS-association domain family 1 (RASSF1), and expression of the RASSF1A isoform of this gene is silenced in tumours by methylation of its promoter. Since methylation is reversible and demethylating agents are currently being used in clinical trials, detection of RASSF1A silencing by promoter hypermethylation has potential clinical uses in cancer diagnosis, prognosis and treatment. RASSF1A belongs to a new family of RAS effectors, of which there are currently 8 members (RASSF1-8). RASSF1-6 each contain a variable N-terminal segment followed by a Ras-association (RA) domain of the Ral-GDS/AF6 type, and a specialised coiled-coil structure known as a SARAH domain extending to the C-terminus. RASSF7-8 contain an N-terminal RA domain and a variable C-terminus. Members of the RASSF family are thought to function as tumour suppressors by regulating the cell cycle and apoptosis. This review will summarise our current knowledge of each member of the RASSF family and in particular what role they play in tumourigenesis, with a special focus on RASSF1A, whose promoter methylation is one of the most frequent alterations found in human tumours.
Figures







References
-
- Downward J. Ras signalling and apoptosis. Curr. Opin. Genet. Dev. 1998;8:49–54. - PubMed
-
- Ponting C.P., Benjamin D.R. A novel family of Ras-binding domains. Trends Biochem. Sci. 1996;21:422–425. - PubMed
-
- Yamamoto T., Taya S., Kaibuchi K. Ras-induced transformation and signaling pathway. J. Biochem. (Tokyo) 1999;126:799–803. - PubMed
-
- Sundaresan V., Ganly P., Hasleton P., Rudd R., Sinha G., Bleehen N.M., Rabbitts P. p53 and chromosome 3 abnormalities, characteristic of malignant lung tumours, are detectable in preinvasive lesions of the bronchus. Oncogene. 1992;7:1989–1997. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

