Characterization of riboflavin (vitamin B2) transport proteins from Bacillus subtilis and Corynebacterium glutamicum
- PMID: 17693491
- PMCID: PMC2168442
- DOI: 10.1128/JB.00590-07
Characterization of riboflavin (vitamin B2) transport proteins from Bacillus subtilis and Corynebacterium glutamicum
Abstract
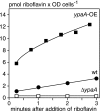
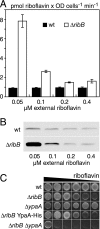
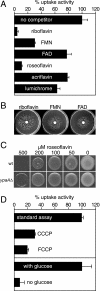
Riboflavin (vitamin B(2)) is the direct precursor of the flavin cofactors flavin mononucleotide and flavin adenine dinucleotide, essential components of cellular biochemistry. In this work we investigated the unrelated proteins YpaA from Bacillus subtilis and PnuX from Corynebacterium glutamicum for a role in riboflavin uptake. Based on the regulation of the corresponding genes by a riboswitch mechanism, both proteins have been predicted to be involved in flavin metabolism. Moreover, their primary structures suggested that these proteins integrate into the cytoplasmic membrane. We provide experimental evidence that YpaA is a plasma membrane protein with five transmembrane domains and a cytoplasmic C terminus. In B. subtilis, riboflavin uptake was increased when ypaA was overexpressed and abolished when ypaA was deleted. Riboflavin uptake activity and the abundance of the YpaA protein were also increased when riboflavin auxotrophic mutants were grown in limiting amounts of riboflavin. YpaA-mediated riboflavin uptake was sensitive to protonophors and reduced in the absence of glucose, demonstrating that the protein requires metabolic energy for substrate translocation. In addition, we demonstrate that PnuX from C. glutamicum also is a riboflavin transporter. Transport by PnuX was not energy dependent and had high apparent affinity for riboflavin (K(m) 11 microM). Roseoflavin, a toxic riboflavin analog, appears to be a substrate of PnuX and YpaA. We propose to designate the gene names ribU for ypaA and ribM for pnuX to reflect that the encoded proteins function in riboflavin uptake and that the genes have different phylogenetic origins.
Figures
Similar articles
-
The physiological role of riboflavin transporter and involvement of FMN-riboswitch in its gene expression in Corynebacterium glutamicum.Appl Microbiol Biotechnol. 2014 May;98(9):4159-68. doi: 10.1007/s00253-014-5570-5. Epub 2014 Feb 16. Appl Microbiol Biotechnol. 2014. PMID: 24531272
-
[Bacillus subtilis ypaA gene regulation mechanism involves FMN-binding sensor RNA].Genetika. 2014 Mar;50(3):364-8. Genetika. 2014. PMID: 25438558 Russian.
-
Engineering of Synechococcus sp. strain PCC 7002 for the photoautotrophic production of light-sensitive riboflavin (vitamin B2).Metab Eng. 2020 Nov;62:275-286. doi: 10.1016/j.ymben.2020.09.010. Epub 2020 Sep 28. Metab Eng. 2020. PMID: 32992032
-
Cell growth and cell division in the rod-shaped actinomycete Corynebacterium glutamicum.Antonie Van Leeuwenhoek. 2008 Jun;94(1):99-109. doi: 10.1007/s10482-008-9224-4. Epub 2008 Feb 19. Antonie Van Leeuwenhoek. 2008. PMID: 18283557 Review.
-
The glucose uptake systems in Corynebacterium glutamicum: a review.World J Microbiol Biotechnol. 2020 Jul 26;36(9):126. doi: 10.1007/s11274-020-02898-z. World J Microbiol Biotechnol. 2020. PMID: 32712859 Review.
Cited by
-
Molecular Elucidation of Riboflavin Production and Regulation in Candida albicans, toward a Novel Antifungal Drug Target.mSphere. 2020 Aug 5;5(4):e00714-20. doi: 10.1128/mSphere.00714-20. mSphere. 2020. PMID: 32759338 Free PMC article.
-
Overview on the Bacterial Iron-Riboflavin Metabolic Axis.Front Microbiol. 2018 Jul 5;9:1478. doi: 10.3389/fmicb.2018.01478. eCollection 2018. Front Microbiol. 2018. PMID: 30026736 Free PMC article. Review.
-
Structure and mechanism of the S component of a bacterial ECF transporter.Nature. 2010 Dec 2;468(7324):717-20. doi: 10.1038/nature09488. Epub 2010 Oct 24. Nature. 2010. PMID: 20972419
-
Archimedes' principle for characterisation of recombinant whole cell biocatalysts.Sci Rep. 2018 Feb 14;8(1):3000. doi: 10.1038/s41598-018-20877-1. Sci Rep. 2018. PMID: 29445212 Free PMC article.
-
Isolation, characterisation and description of the roseoflavin producer Streptomyces berlinensis sp. nov.Environ Microbiol Rep. 2024 Apr;16(2):e13266. doi: 10.1111/1758-2229.13266. Environ Microbiol Rep. 2024. PMID: 38653477 Free PMC article.
References
-
- Bacher, A., S. Eberhardt, M. Fischer, K. Kis, and G. Richter. 2000. Biosynthesis of vitamin B2 (riboflavin). Annu. Rev. Nutr. 20:153-167. - PubMed
-
- Bandrin, S. V., P. M. Rabinovich, and A. I. Stepanov. 1993. Three linkage groups involved in riboflavin biosynthesis in E. coli. Sovi. Genet. 19:1103-1109. - PubMed
-
- Banerjee, R., and A. Batschauer. 2005. Plant blue-light receptors. Planta 220:498-502. - PubMed
-
- Cecchini, G., M. Perl, J. Lipsick, T. P. Singer, and E. B. Kearney. 1979. Transport and binding of riboflavin by Bacillus subtilis. J. Biol. Chem. 254:7295-7301. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

