Characterization of an acid-dependent arginine decarboxylase enzyme from Chlamydophila pneumoniae
- PMID: 17693492
- PMCID: PMC2168457
- DOI: 10.1128/JB.00772-07
Characterization of an acid-dependent arginine decarboxylase enzyme from Chlamydophila pneumoniae
Abstract
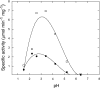
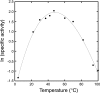
Genome sequences from members of the Chlamydiales encode diverged homologs of a pyruvoyl-dependent arginine decarboxylase enzyme that nonpathogenic euryarchaea use in polyamine biosynthesis. The Chlamydiales lack subsequent genes required for polyamine biosynthesis and probably obtain polyamines from their host cells. To identify the function of this protein, the CPn1032 homolog from the respiratory pathogen Chlamydophila pneumoniae was heterologously expressed and purified. This protein self-cleaved to form a reactive pyruvoyl group, and the subunits assembled into a thermostable (alphabeta)(3) complex. The mature enzyme specifically catalyzed the decarboxylation of L-arginine, with an unusually low pH optimum of 3.4. The CPn1032 gene complemented a mutation in the Escherichia coli adiA gene, which encodes a pyridoxal 5'-phosphate-dependent arginine decarboxylase, restoring arginine-dependent acid resistance. Acting together with a putative arginine-agmatine antiporter, the CPn1032 homologs may have evolved convergently to form an arginine-dependent acid resistance system. These genes are the first evidence that obligately intracellular chlamydiae may encounter acidic conditions. Alternatively, this system could reduce the host cell arginine concentration and produce inhibitors of nitric oxide synthase.
Figures




Similar articles
-
Outer and inner membrane proteins compose an arginine-agmatine exchange system in Chlamydophila pneumoniae.J Bacteriol. 2008 Nov;190(22):7431-40. doi: 10.1128/JB.00652-08. Epub 2008 Sep 12. J Bacteriol. 2008. PMID: 18790867 Free PMC article.
-
Methanococcus jannaschii uses a pyruvoyl-dependent arginine decarboxylase in polyamine biosynthesis.J Biol Chem. 2002 Jun 28;277(26):23500-7. doi: 10.1074/jbc.M203467200. Epub 2002 Apr 29. J Biol Chem. 2002. PMID: 11980912
-
Purification and characterization of a 4-hydroxybenzoate decarboxylase from Chlamydophila pneumoniae AR39.Curr Microbiol. 2007 Feb;54(2):102-7. doi: 10.1007/s00284-006-0153-z. Epub 2007 Jan 5. Curr Microbiol. 2007. PMID: 17211544
-
Regulation of polyamine biosynthesis by antizyme and some recent developments relating the induction of polyamine biosynthesis to cell growth. Review.Biosci Rep. 1985 Mar;5(3):189-204. doi: 10.1007/BF01119588. Biosci Rep. 1985. PMID: 3893559 Review.
-
Agmatine: at the crossroads of the arginine pathways.Ann N Y Acad Sci. 2003 Dec;1009:34-43. doi: 10.1196/annals.1304.004. Ann N Y Acad Sci. 2003. PMID: 15028568 Review.
Cited by
-
Crenarchaeal arginine decarboxylase evolved from an S-adenosylmethionine decarboxylase enzyme.J Biol Chem. 2008 Sep 19;283(38):25829-38. doi: 10.1074/jbc.M802674200. Epub 2008 Jul 23. J Biol Chem. 2008. PMID: 18650422 Free PMC article.
-
Improved acid stress survival of Lactococcus lactis expressing the histidine decarboxylation pathway of Streptococcus thermophilus CHCC1524.J Biol Chem. 2012 Mar 30;287(14):11195-204. doi: 10.1074/jbc.M111.330704. Epub 2012 Feb 17. J Biol Chem. 2012. PMID: 22351775 Free PMC article.
-
Genome economization in the endosymbiont of the wood roach Cryptocercus punctulatus due to drastic loss of amino acid synthesis capabilities.Genome Biol Evol. 2011;3:1437-48. doi: 10.1093/gbe/evr118. Epub 2011 Nov 16. Genome Biol Evol. 2011. PMID: 22094859 Free PMC article.
-
Vaginal biogenic amines: biomarkers of bacterial vaginosis or precursors to vaginal dysbiosis?Front Physiol. 2015 Sep 29;6:253. doi: 10.3389/fphys.2015.00253. eCollection 2015. Front Physiol. 2015. PMID: 26483694 Free PMC article.
-
From Protease to Decarboxylase: THE MOLECULAR METAMORPHOSIS OF PHOSPHATIDYLSERINE DECARBOXYLASE.J Biol Chem. 2015 Apr 24;290(17):10972-80. doi: 10.1074/jbc.M115.642413. Epub 2015 Feb 26. J Biol Chem. 2015. PMID: 25724650 Free PMC article.
References
-
- Al-Younes, H. M., J. Gussmann, P. R. Braun, V. Brinkmann, and T. F. Meyer. 2006. Naturally occurring amino acids differentially influence the development of Chlamydia trachomatis and Chlamydia (Chlamydophila) pneumoniae. J. Med. Microbiol. 55:879-886. - PubMed
-
- Bavoil, P. M., R.-C. Hsia, and D. M. Ojcius. 2000. Closing in on Chlamydia and its intracellular bag of tricks. Microbiology 146:2723-2731. - PubMed
-
- Blethen, S. L., E. A. Boeker, and E. E. Snell. 1968. Arginine decarboxylase from Escherichia coli. I. Purification and specificity for substrates and coenzyme. J. Biol. Chem. 243:1671-1677. - PubMed
-
- Carratelli, C. R., A. Rizzo, R. Paolillo, M. R. Catania, P. Catalanotti, and F. Rossano. 2005. Effect of nitric oxide on the growth of Chlamydophila pneumoniae. Can. J. Microbiol. 51:941-947. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

