Quorum-sensing regulation of the production of Blp bacteriocins in Streptococcus thermophilus
- PMID: 17693498
- PMCID: PMC2168441
- DOI: 10.1128/JB.00966-07
Quorum-sensing regulation of the production of Blp bacteriocins in Streptococcus thermophilus
Abstract
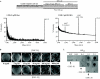
The blp gene cluster identified in the genome sequences of Streptococcus thermophilus (blp(St)) LMG18311, CNRZ1066, and LMD-9 displays all the characteristics of a class II bacteriocin locus. In the present study, we showed that the blp(St) locus is only fully functional in strain LMD-9 and regulates the production of antimicrobial peptides that inhibit strains LMG18311 and CNRZ1066. The blp(St) cluster of LMD-9 contains 23 genes that are transcriptionally organized in six operons: blpABC(St) (peptide transporter genes and pheromone gene); blpRH(St) (two-component regulatory system genes); blpD(St)-orf1, blpU(St)-orf3, and blpE-F(St) (bacteriocin precursors and immunity genes); and blpG-X(St) (unknown function). All the operons, except the regulatory unit blpRH(St), were shown to be coregulated at the transcriptional level by a quorum-sensing mechanism involving the mature S. thermophilus pheromone BlpC* (BlpC*(St)), which was extracellularly detected as two active forms (30 and 19 amino acids). These operons are differentially transcribed depending on growth phase and pheromone concentration. They all contain a motif with two imperfect direct repeats in their mapped promoter regions that could serve as binding sites of the response regulator BlpR(St). Through the construction of deletion mutants, the blp(St) locus of strain LMD-9 was shown to encode all the essential functions associated with bacteriocin production, quorum-sensing regulation, and immunity.
Figures

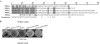


References
-
- Aktypis, A., and G. Kalantzopoulos. 2003. Purification and characterization of thermophilin ST-1, a novel bacteriocin produced by Streptococcus thermophilus ACA-DC 0001. Lait 83:365-378. - PubMed
-
- Aktypis, A., G. Kalantzopoulos, J. H. J. Huis in't Veld, and B. ten Brink. 1998. Purification and characterization of thermophilin T, a novel bacteriocin produced by Streptococcus thermophilus ACA-DC 0040. J. Appl. Microbiol. 84:568-576. - PubMed
-
- Andrade, J. M., F. Cairrao, and C. M. Arraiano. 2006. RNase R affects gene expression in stationary phase: regulation of ompA. Mol. Microbiol. 60:219-228. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

