Proteins P24 and P41 function in the regulation of terminal-organelle development and gliding motility in Mycoplasma pneumoniae
- PMID: 17693502
- PMCID: PMC2168445
- DOI: 10.1128/JB.00867-07
Proteins P24 and P41 function in the regulation of terminal-organelle development and gliding motility in Mycoplasma pneumoniae
Abstract
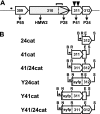
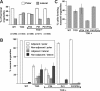
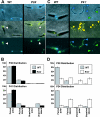
Mycoplasma pneumoniae is a major cause of bronchitis and atypical pneumonia in humans. This cell wall-less bacterium has a complex terminal organelle that functions in cytadherence and gliding motility. The gliding mechanism is unknown but is coordinated with terminal-organelle development during cell division. Disruption of M. pneumoniae open reading frame MPN311 results in loss of protein P41 and downstream gene product P24. P41 localizes to the base of the terminal organelle and is required to anchor the terminal organelle to the cell body, but during cell division, MPN311 insertion mutants also fail to properly regulate nascent terminal-organelle development spatially or gliding activity temporally. We measured gliding velocity and frequency and used fluorescent protein fusions and time-lapse imaging to assess the roles of P41 and P24 individually in terminal-organelle development and gliding function. P41 was necessary for normal gliding velocity and proper spatial positioning of new terminal organelles, while P24 was required for gliding frequency and new terminal-organelle formation at wild-type rates. However, P41 was essential for P24 function, and in the absence of P41, P24 exhibited a dynamic localization pattern. Finally, protein P28 requires P41 for stability, but analysis of a P28(-) mutant established that the MPN311 mutant phenotype was not a function of loss of P28.
Figures



Similar articles
-
Mycoplasma pneumoniae J-domain protein required for terminal organelle function.Mol Microbiol. 2009 Mar;71(5):1296-307. doi: 10.1111/j.1365-2958.2009.06602.x. Epub 2009 Jan 29. Mol Microbiol. 2009. PMID: 19183275 Free PMC article.
-
Cytoskeletal protein P41 is required to anchor the terminal organelle of the wall-less prokaryote Mycoplasma pneumoniae.Mol Microbiol. 2007 Jan;63(1):44-53. doi: 10.1111/j.1365-2958.2006.05507.x. Epub 2006 Dec 5. Mol Microbiol. 2007. PMID: 17163973
-
Use of fluorescent-protein tagging to determine the subcellular localization of mycoplasma pneumoniae proteins encoded by the cytadherence regulatory locus.J Bacteriol. 2004 Oct;186(20):6944-55. doi: 10.1128/JB.186.20.6944-6955.2004. J Bacteriol. 2004. PMID: 15466048 Free PMC article.
-
Structure, function, and assembly of the terminal organelle of Mycoplasma pneumoniae.FEMS Microbiol Lett. 2001 Apr 20;198(1):1-7. doi: 10.1111/j.1574-6968.2001.tb10610.x. FEMS Microbiol Lett. 2001. PMID: 11325545 Review.
-
Mycoplasma pneumoniae cytadherence: organization and assembly of the attachment organelle.Trends Microbiol. 1998 Jan;6(1):15-8. doi: 10.1016/S0966-842X(97)01168-2. Trends Microbiol. 1998. PMID: 9481818 Review.
Cited by
-
Mycoplasma pneumoniae Infections: Pathogenesis and Vaccine Development.Pathogens. 2021 Jan 25;10(2):119. doi: 10.3390/pathogens10020119. Pathogens. 2021. PMID: 33503845 Free PMC article. Review.
-
Mycoplasma pneumoniae J-domain protein required for terminal organelle function.Mol Microbiol. 2009 Mar;71(5):1296-307. doi: 10.1111/j.1365-2958.2009.06602.x. Epub 2009 Jan 29. Mol Microbiol. 2009. PMID: 19183275 Free PMC article.
-
First identification of proteins involved in motility of Mycoplasma gallisepticum.Vet Res. 2014 Oct 17;45(1):99. doi: 10.1186/s13567-014-0099-2. Vet Res. 2014. PMID: 25323771 Free PMC article.
-
Functional domain analysis of the Mycoplasma pneumoniae co-chaperone TopJ.Mol Microbiol. 2010 Jul 1;77(1):158-69. doi: 10.1111/j.1365-2958.2010.07196.x. Epub 2010 May 12. Mol Microbiol. 2010. PMID: 20487283 Free PMC article.
-
P40 and P90 from Mpn142 are Targets of Multiple Processing Events on the Surface of Mycoplasma pneumoniae.Proteomes. 2015 Dec 16;3(4):512-537. doi: 10.3390/proteomes3040512. Proteomes. 2015. PMID: 28248283 Free PMC article.
References
-
- Balish, M. F., S. M. Ross, M. Fisseha, and D. C. Krause. 2003. Deletion analysis identifies key functional domains of the cytadherence-associated protein HMW2 of Mycoplasma pneumoniae. Mol. Microbiol. 50:1507-1516. - PubMed
-
- Bredt, W. 1968. Motility and multiplication of Mycoplasma pneumoniae. A phase contrast study. Pathol. Microbiol. (Basel) 32:321-326. - PubMed
-
- Dandekar, T., M. Huynen, J. T. Regula, B. Ueberle, C. U. Zimmermann, M. A. Andrade, T. Doerks, L. Sanchez-Pulido, B. Snel, M. Suyama, Y. P. Yuan, R. Herrmann, and P. Bork. 2000. Re-annotating the Mycoplasma pneumoniae genome sequence: adding value, function and reading frames. Nucleic Acids Res. 28:3278-3288. - PMC - PubMed
-
- Gitai, Z., M. Thanbichler, and L. Shapiro. 2005. The choreographed dynamics of bacterial chromosomes. Trends Microbiol. 13:221-228. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

