A molecular timetable for apical bud formation and dormancy induction in poplar
- PMID: 17693531
- PMCID: PMC2002631
- DOI: 10.1105/tpc.107.052811
A molecular timetable for apical bud formation and dormancy induction in poplar
Abstract
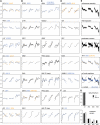
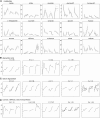
The growth of perennial plants in the temperate zone alternates with periods of dormancy that are typically initiated during bud development in autumn. In a systems biology approach to unravel the underlying molecular program of apical bud development in poplar (Populus tremula x Populus alba), combined transcript and metabolite profiling were applied to a high-resolution time course from short-day induction to complete dormancy. Metabolite and gene expression dynamics were used to reconstruct the temporal sequence of events during bud development. Importantly, bud development could be dissected into bud formation, acclimation to dehydration and cold, and dormancy. To each of these processes, specific sets of regulatory and marker genes and metabolites are associated and provide a reference frame for future functional studies. Light, ethylene, and abscisic acid signal transduction pathways consecutively control bud development by setting, modifying, or terminating these processes. Ethylene signal transduction is positioned temporally between light and abscisic acid signals and is putatively activated by transiently low hexose pools. The timing and place of cell proliferation arrest (related to dormancy) and of the accumulation of storage compounds (related to acclimation processes) were established within the bud by electron microscopy. Finally, the identification of a large set of genes commonly expressed during the growth-to-dormancy transitions in poplar apical buds, cambium, or Arabidopsis thaliana seeds suggests parallels in the underlying molecular mechanisms in different plant organs.
Figures







Similar articles
-
Gene expression during the induction, maintenance, and release of dormancy in apical buds of poplar.J Exp Bot. 2007;58(15-16):4047-60. doi: 10.1093/jxb/erm261. Epub 2007 Nov 26. J Exp Bot. 2007. PMID: 18039739
-
Overexpression of DEMETER, a DNA demethylase, promotes early apical bud maturation in poplar.Plant Cell Environ. 2017 Nov;40(11):2806-2819. doi: 10.1111/pce.13056. Epub 2017 Oct 12. Plant Cell Environ. 2017. PMID: 28810288
-
CENL1 expression in the rib meristem affects stem elongation and the transition to dormancy in Populus.Plant Cell. 2008 Jan;20(1):59-74. doi: 10.1105/tpc.107.056721. Epub 2008 Jan 11. Plant Cell. 2008. PMID: 18192437 Free PMC article.
-
The embryonic shoot: a lifeline through winter.J Exp Bot. 2014 Apr;65(7):1699-712. doi: 10.1093/jxb/ert413. Epub 2013 Dec 24. J Exp Bot. 2014. PMID: 24368502 Review.
-
Interaction of Phytohormones and External Environmental Factors in the Regulation of the Bud Dormancy in Woody Plants.Int J Mol Sci. 2023 Dec 6;24(24):17200. doi: 10.3390/ijms242417200. Int J Mol Sci. 2023. PMID: 38139028 Free PMC article. Review.
Cited by
-
BRANCHED1 promotes axillary bud dormancy in response to shade in Arabidopsis.Plant Cell. 2013 Mar;25(3):834-50. doi: 10.1105/tpc.112.108480. Epub 2013 Mar 22. Plant Cell. 2013. PMID: 23524661 Free PMC article.
-
Metabolomics analysis reveals Embden Meyerhof Parnas pathway activation and flavonoids accumulation during dormancy transition in tree peony.BMC Plant Biol. 2020 Oct 23;20(1):484. doi: 10.1186/s12870-020-02692-x. BMC Plant Biol. 2020. PMID: 33096979 Free PMC article.
-
Dual role of tree florigen activation complex component FD in photoperiodic growth control and adaptive response pathways.Proc Natl Acad Sci U S A. 2015 Mar 10;112(10):3140-5. doi: 10.1073/pnas.1423440112. Epub 2015 Feb 23. Proc Natl Acad Sci U S A. 2015. PMID: 25713384 Free PMC article.
-
Time-coursed transcriptome analysis identifies key expressional regulation in growth cessation and dormancy induced by short days in Paulownia.Sci Rep. 2019 Nov 12;9(1):16602. doi: 10.1038/s41598-019-53283-2. Sci Rep. 2019. PMID: 31719639 Free PMC article.
-
Unravelling the Role of Epigenetic Modifications in Development and Reproduction of Angiosperms: A Critical Appraisal.Front Genet. 2022 May 18;13:819941. doi: 10.3389/fgene.2022.819941. eCollection 2022. Front Genet. 2022. PMID: 35664328 Free PMC article. Review.
References
-
- Arend, M., and Fromm, J. (2003). Ultrastructural changes in cambial cell derivatives during xylem differentiation in poplar. Plant Biol. 5 255–264.
-
- Barrero, J.M., Rodríguez, P.L., Quesada, V., Piqueras, P., Ponce, M.R., and Micol, J.L. (2006). Both abscisic acid (ABA)-dependent and ABA-independent pathways govern the induction of NCED3, AAO3 and ABA1 in response to salt stress. Plant Cell Environ. 29 2000–2008. - PubMed
-
- Baumbusch, L.O., Hughes, D.W., Galau, G.A., and Jakobsen, K.S. (2004). LEC1, FUS3, ABI3 and Em expression reveals no correlation with dormancy in Arabidopsis. J. Exp. Bot. 55 77–87. - PubMed
-
- Beemster, G.T.S., De Veylder, L., Vercruysse, S., West, G., Rombaut, D., Van Hummelen, P., Galichet, A., Gruissem, W., Inzé, D., and Vuylsteke, M. (2005). Genome-wide analysis of gene expression profiles associated with cell cycle transitions in growing organs of Arabidopsis. Plant Physiol. 138 734–743. - PMC - PubMed
-
- Benedict, C., Skinner, J.S., Meng, R., Chang, Y., Bhalerao, R., Huner, N.P.A., Finn, C.E., Chen, T.H.H., and Hurry, V. (2006). The CBF1-dependent low temperature signalling pathway, regulon and increase in freeze tolerance are conserved in Populus spp. Plant Cell Environ. 29 1259–1272. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

