Signaling from an altered cell wall to the nucleus mediates sugar-responsive growth and development in Arabidopsis thaliana
- PMID: 17693536
- PMCID: PMC2002624
- DOI: 10.1105/tpc.106.049965
Signaling from an altered cell wall to the nucleus mediates sugar-responsive growth and development in Arabidopsis thaliana
Abstract
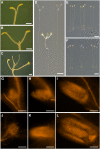
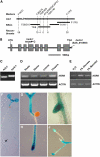
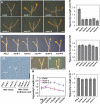
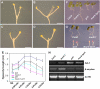
Sugars such as glucose function as signal molecules that regulate gene expression, growth, and development in plants, animals, and yeast. To understand the molecular mechanisms of sugar responses, we isolated and characterized an Arabidopsis thaliana mutant, high sugar response8 (hsr8), which enhances sugar-responsive growth and gene expression. Light-grown hsr8 plants exhibited increased starch and anthocyanin and reduced chlorophyll content in response to glucose treatment. Dark-grown hsr8 seedlings showed glucose-hypersensitive hypocotyl elongation and development. The HSR8 gene, isolated using map-based cloning, was allelic to the MURUS4 (MUR4) gene involved in arabinose synthesis. Dark-grown mur1 and mur3 seedlings also exhibited similar sugar responses to hsr8/mur4. The sugar-hypersensitive phenotypes of hsr8/mur4, mur1, and mur3 were rescued by boric acid, suggesting that alterations in the cell wall cause hypersensitive sugar-responsive phenotypes. Genetic analysis showed that sugar-hypersensitive responses in hsr8 mutants were suppressed by pleiotropic regulatory locus1 (prl1), indicating that nucleus-localized PRL1 is required for enhanced sugar responses in hsr8 mutant plants. Microarray analysis revealed that the expression of many cell wall-related and sugar-responsive genes was altered in mur4-1, and the expression of a significant proportion of these genes was restored to wild-type levels in the mur4-1 prl1 double mutant. These findings reveal a pathway that signals changes in the cell wall through PRL1 to altered gene expression and sugar-responsive metabolic, growth, and developmental changes.
Figures



Similar articles
-
TANG1, Encoding a Symplekin_C Domain-Contained Protein, Influences Sugar Responses in Arabidopsis.Plant Physiol. 2015 Jul;168(3):1000-12. doi: 10.1104/pp.15.00288. Epub 2015 May 22. Plant Physiol. 2015. PMID: 26002908 Free PMC article.
-
The Mediator complex subunits MED25/PFT1 and MED8 are required for transcriptional responses to changes in cell wall arabinose composition and glucose treatment in Arabidopsis thaliana.BMC Plant Biol. 2015 Sep 5;15:215. doi: 10.1186/s12870-015-0592-4. BMC Plant Biol. 2015. PMID: 26341899 Free PMC article.
-
GABA accumulation causes cell elongation defects and a decrease in expression of genes encoding secreted and cell wall-related proteins in Arabidopsis thaliana.Plant Cell Physiol. 2011 May;52(5):894-908. doi: 10.1093/pcp/pcr041. Epub 2011 Apr 6. Plant Cell Physiol. 2011. PMID: 21471118 Free PMC article.
-
Arabinose biosynthesis is critical for salt stress tolerance in Arabidopsis.New Phytol. 2019 Oct;224(1):274-290. doi: 10.1111/nph.15867. Epub 2019 May 17. New Phytol. 2019. PMID: 31009077
-
Arabidopsis constitutive photomorphogenic mutant, bls1, displays altered brassinosteroid response and sugar sensitivity.Plant Mol Biol. 2004 Sep;56(2):185-201. doi: 10.1007/s11103-004-2799-x. Plant Mol Biol. 2004. PMID: 15604737
Cited by
-
Discover and connect cellular signaling.Plant Physiol. 2010 Oct;154(2):562-6. doi: 10.1104/pp.110.161364. Plant Physiol. 2010. PMID: 20921185 Free PMC article. No abstract available.
-
Identification of an EMS-induced causal mutation in a gene required for boron-mediated root development by low-coverage genome re-sequencing in Arabidopsis.Plant Signal Behav. 2013 Jan;8(1):e22534. doi: 10.4161/psb.22534. Epub 2012 Oct 26. Plant Signal Behav. 2013. PMID: 23104114 Free PMC article.
-
Cell wall integrity: targeted post-synthetic modifications to reveal its role in plant growth and defense against pathogens.Plant Signal Behav. 2013 Sep;8(9):e25435. doi: 10.4161/psb.25435. Epub 2013 Jun 20. Plant Signal Behav. 2013. PMID: 23857352 Free PMC article. Review.
-
Developmental consequences of the tumorous shoot development1 mutation, a novel allele of the cellulose-synthesizing KORRIGAN1 gene.Plant Mol Biol. 2009 Dec;71(6):641-55. doi: 10.1007/s11103-009-9546-2. Epub 2009 Oct 14. Plant Mol Biol. 2009. PMID: 19826767
-
Dolichol biosynthesis and its effects on the unfolded protein response and abiotic stress resistance in Arabidopsis.Plant Cell. 2008 Jul;20(7):1879-98. doi: 10.1105/tpc.108.061150. Epub 2008 Jul 8. Plant Cell. 2008. PMID: 18612099 Free PMC article.
References
-
- Alonso, J.M., Hirayama, T., Roman, G., Nourizadeh, S., and Ecker, J.R. (1999). EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 284 2148–2152. - PubMed
-
- Arioli, T., et al. (1998). Molecular analysis of cellulose biosynthesis in Arabidopsis. Science 279 717–720. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

