Mixed infections, cryptic diversity, and vector-borne pathogens: evidence from Polygenis fleas and Bartonella species
- PMID: 17693558
- PMCID: PMC2075021
- DOI: 10.1128/AEM.00228-07
Mixed infections, cryptic diversity, and vector-borne pathogens: evidence from Polygenis fleas and Bartonella species
Abstract
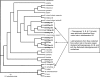
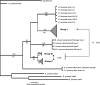
Coinfections within hosts present opportunities for horizontal gene transfer between strains and competitive interactions between genotypes and thus can be a critical element of the lifestyles of pathogens. Bartonella spp. are Alphaproteobacteria that parasitize mammalian erythrocytes and endothelial cells. Their vectors are thought to be various biting arthropods, such as fleas, ticks, mites, and lice, and they are commonly cited as agents of various emerging diseases. Coinfections by different Bartonella strains and species can be common in mammals, but little is known about specificity and coinfections in arthropod vectors. We surveyed the rate of mixed infections of Bartonella in flea vectors (Polygenis gwyni) parasitizing cotton rats (Sigmodon hispidus) in which previous surveys indicated high rates of infection. We found that nearly all fleas (20 of 21) harbored one or more strains of Bartonella, with rates of coinfection approaching 90%. A strain previously identified as common in cotton rats was also common in their fleas. However, another common strain in cotton rats was absent from P. gwyni, while a rare cotton rat strain was quite common in P. gwyni. Surprisingly, some samples were also coinfected with a strain phylogenetically related to Bartonella clarridgeiae, which is typically associated with felids and ruminants. Finally, a locus (pap31) that is characteristically borne on phage in Bartonella was successfully sequenced from most samples. However, sequence diversity in pap31 was novel in the P. gwyni samples, relative to other Bartonella previously typed with pap31, emphasizing the likelihood of large reservoirs of cryptic diversity in natural populations of the pathogen.
Figures
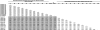
References
-
- Alexander, B. 1995. A review of bartonellosis in Ecuador and Colombia. Am. J. Trop. Med. Hyg. 52:354-359. - PubMed
-
- Alsmark, C. M., A. C. Frank, E. O. Karlberg, B. A. Legault, D. H. Ardell, B. Canback, A. S. Eriksson, A. K. Naslund, S. A. Handley, M. Huvet, B. La Scola, M. Holmberg, and S. G. E. Andersson. 2004. The louse-borne human pathogen Bartonella quintana is a genomic derivative of the zoonotic agent Bartonella henselae. Proc. Acad. Natl. Sci. USA 101:9716-9721. - PMC - PubMed
-
- Barbet, A. F., P. F. M. Meeus, M. Belanger, M. V. Bowie, J. Yi, A. M. Lundgren, A. R. Alleman, S. J. Wong, F. K. Chu, U. G. Munderloh, and S. D. Jauron. 2003. Expression of multiple outer membrane protein sequence variants from a single genomic locus of Anaplasma phagocytophilum. Infect. Immun. 71:1706-1718. - PMC - PubMed
-
- Boucher, Y., C. J. Douady, R. T. Papke, D. A. Walsh, M. E. R. Boudreau, C. L. Nesbo, R. J. Case, and W. F. Doolittle. 2003. Lateral gene transfer and the origins of prokaryotic groups. Annu. Rev. Genet. 37:283-328. - PubMed
-
- Boulouis, H. J., C. C. Chang, J. B. Henn, R. W. Kasten, and B. B. Chomel. 2005. Factors associated with the rapid emergence of zoonotic Bartonella infections. Vet. Res. 36:383-410. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Miscellaneous

