FR3D: finding local and composite recurrent structural motifs in RNA 3D structures
- PMID: 17694311
- PMCID: PMC2837920
- DOI: 10.1007/s00285-007-0110-x
FR3D: finding local and composite recurrent structural motifs in RNA 3D structures
Abstract
New methods are described for finding recurrent three-dimensional (3D) motifs in RNA atomic-resolution structures. Recurrent RNA 3D motifs are sets of RNA nucleotides with similar spatial arrangements. They can be local or composite. Local motifs comprise nucleotides that occur in the same hairpin or internal loop. Composite motifs comprise nucleotides belonging to three or more different RNA strand segments or molecules. We use a base-centered approach to construct efficient, yet exhaustive search procedures using geometric, symbolic, or mixed representations of RNA structure that we implement in a suite of MATLAB programs, "Find RNA 3D" (FR3D). The first modules of FR3D preprocess structure files to classify base-pair and -stacking interactions. Each base is represented geometrically by the position of its glycosidic nitrogen in 3D space and by the rotation matrix that describes its orientation with respect to a common frame. Base-pairing and base-stacking interactions are calculated from the base geometries and are represented symbolically according to the Leontis/Westhof basepairing classification, extended to include base-stacking. These data are stored and used to organize motif searches. For geometric searches, the user supplies the 3D structure of a query motif which FR3D uses to find and score geometrically similar candidate motifs, without regard to the sequential position of their nucleotides in the RNA chain or the identity of their bases. To score and rank candidate motifs, FR3D calculates a geometric discrepancy by rigidly rotating candidates to align optimally with the query motif and then comparing the relative orientations of the corresponding bases in the query and candidate motifs. Given the growing size of the RNA structure database, it is impossible to explicitly compute the discrepancy for all conceivable candidate motifs, even for motifs with less than ten nucleotides. The screening algorithm that we describe finds all candidate motifs whose geometric discrepancy with respect to the query motif falls below a user-specified cutoff discrepancy. This technique can be applied to RMSD searches. Candidate motifs identified geometrically may be further screened symbolically to identify those that contain particular basepair types or base-stacking arrangements or that conform to sequence continuity or nucleotide identity constraints. Purely symbolic searches for motifs containing user-defined sequence, continuity and interaction constraints have also been implemented. We demonstrate that FR3D finds all occurrences, both local and composite and with nucleotide substitutions, of sarcin/ricin and kink-turn motifs in the 23S and 5S ribosomal RNA 3D structures of the H. marismortui 50S ribosomal subunit and assigns the lowest discrepancy scores to bona fide examples of these motifs. The search algorithms have been optimized for speed to allow users to search the non-redundant RNA 3D structure database on a personal computer in a matter of minutes.
Figures









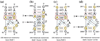
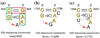

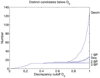


Similar articles
-
WebFR3D--a server for finding, aligning and analyzing recurrent RNA 3D motifs.Nucleic Acids Res. 2011 Jul;39(Web Server issue):W50-5. doi: 10.1093/nar/gkr249. Epub 2011 Apr 22. Nucleic Acids Res. 2011. PMID: 21515634 Free PMC article.
-
Automated classification of RNA 3D motifs and the RNA 3D Motif Atlas.RNA. 2013 Oct;19(10):1327-40. doi: 10.1261/rna.039438.113. Epub 2013 Aug 22. RNA. 2013. PMID: 23970545 Free PMC article.
-
RNAMotifScan: automatic identification of RNA structural motifs using secondary structural alignment.Nucleic Acids Res. 2010 Oct;38(18):e176. doi: 10.1093/nar/gkq672. Epub 2010 Aug 8. Nucleic Acids Res. 2010. PMID: 20696653 Free PMC article.
-
The RNA 3D Motif Atlas: Computational methods for extraction, organization and evaluation of RNA motifs.Methods. 2016 Jul 1;103:99-119. doi: 10.1016/j.ymeth.2016.04.025. Epub 2016 Apr 25. Methods. 2016. PMID: 27125735 Free PMC article. Review.
-
The building blocks and motifs of RNA architecture.Curr Opin Struct Biol. 2006 Jun;16(3):279-87. doi: 10.1016/j.sbi.2006.05.009. Epub 2006 May 19. Curr Opin Struct Biol. 2006. PMID: 16713707 Free PMC article. Review.
Cited by
-
DETECTING CONFORMATIONAL DIFFERENCES BETWEEN RNA 3D STRUCTURES.JP J Biostat. 2015 Dec;12(2):99-115. doi: 10.17654/JPJBDec2015_099_115. Epub 2016 Jan 1. JP J Biostat. 2015. PMID: 27330250 Free PMC article.
-
On the predictibility of A-minor motifs from their local contexts.RNA Biol. 2022 Jan;19(1):1208-1227. doi: 10.1080/15476286.2022.2144611. RNA Biol. 2022. PMID: 36384383 Free PMC article.
-
Towards 3D structure prediction of large RNA molecules: an integer programming framework to insert local 3D motifs in RNA secondary structure.Bioinformatics. 2012 Jun 15;28(12):i207-14. doi: 10.1093/bioinformatics/bts226. Bioinformatics. 2012. PMID: 22689763 Free PMC article.
-
Predicting RNA 3D structure using a coarse-grain helix-centered model.RNA. 2015 Jun;21(6):1110-21. doi: 10.1261/rna.047522.114. Epub 2015 Apr 22. RNA. 2015. PMID: 25904133 Free PMC article.
-
RNANet: an automatically built dual-source dataset integrating homologous sequences and RNA structures.Bioinformatics. 2021 Jun 9;37(9):1218-1224. doi: 10.1093/bioinformatics/btaa944. Bioinformatics. 2021. PMID: 33135044 Free PMC article.
References
-
- Adams PL, Stahley MR, Kosek AB, Wang J, Strobel SA. Crystal structure of a self-splicing group I intron with both exons. Nature. 2004;430(6995):45–50. - PubMed
-
- Babcock MS, Pednaul TEP, Olson WK. Nucleic acid structure analysis. mathematics for local Cartesian and helical structure parameters that are truly comparable between structures. J. Mol. Biol. 1994;237(1):125–156. - PubMed
-
- Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289(5481):905–920. - PubMed
-
- Bayley MJ, Gardiner EJ, Willett P, Artymiuk PJ. A fourier fingerprint-based method for protein surface representation. J. Chem. Inf. Model. 2005;45(3):696–707. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

