Characterisation of the expression and function of the GM-CSF receptor alpha-chain in mice
- PMID: 17694571
- PMCID: PMC2699419
- DOI: 10.1002/eji.200636892
Characterisation of the expression and function of the GM-CSF receptor alpha-chain in mice
Abstract
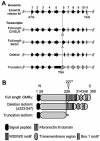
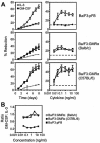
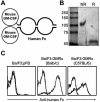
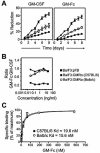
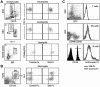
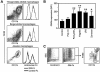
The granulocyte-macrophage colony-stimulating factor (GM-CSF) is a hematopoietic cytokine able to regulate a variety of cell functions including differentiation of macrophages and granulocytes, dendritic cell development and the maintenance of homeostasis. It binds specifically to its receptor, which is composed of a cytokine-specific alpha-chain (GM-CSF receptor alpha-chain, GMRalpha) and a beta-chain shared with the receptors for interleukin-3 and interleukin-5. In this report, we present a comprehensive study of GMRalpha in the mouse. We have found that the mouse GMRalpha is polymorphic and alternatively spliced. In the absence of specific antibodies, we generated a novel chimeric protein containing the Fc fragment of human IgG1 coupled to mouse GM-CSF, which was able to specifically bind to GMRalpha and induce proliferation of GMRalpha-transduced Ba/F3 cells. We used this reagent to perform the first detailed FACS study of the surface expression of mouse GMRalpha by leucocytes. Highest expression was found on monocytes and granulocytes, and variable expression on tissue macrophages. The GM-CSF receptor in mice is specifically expressed by myeloid cells and is useful for the detection of novel uncharacterised myeloid populations. The ability to detect GM-CSF receptor expression in experimental studies should greatly facilitate the analysis of its role in immune pathologies.
Figures

References
-
- Hamilton JA, Anderson GP. GM-CSF biology. Growth Factors. 2004;22:225–231. - PubMed
-
- Armitage JO. Emerging applications of recombinant human granulocyte-macrophage colony-stimulating factor. Blood. 1998;92:4491–4508. - PubMed
-
- Trapnell BC, Whitsett JA. GM-CSF regulates pulmonary surfactant homeostasis and alveolar macrophage-mediated innate host defense. Annu. Rev. Physiol. 2002;64:775–802. - PubMed
-
- Reed JAH, Rice WR, Zsengeller ZK, Wert SE, Dranoff G, Whitsett JA. GM-CSF enhances lung growth and causes alveolar type II epithelial cell hyperplasia in transgenic mice. Am. J. Physiol. Lung Cell. Mol. Physiol. 1997;273:L715–L725. - PubMed
-
- Wognum AW, Westerman Y, Visser TP, Wagemaker G. Distribution of receptors for granulocyte-macrophage colony-stimulating factor on immature CD34+ bone marrow cells, differentiating monomyeloid progenitors, and mature blood cell subsets. Blood. 1994;84:764–774. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

