The role of AtMUS81 in interference-insensitive crossovers in A. thaliana
- PMID: 17696612
- PMCID: PMC1941751
- DOI: 10.1371/journal.pgen.0030132
The role of AtMUS81 in interference-insensitive crossovers in A. thaliana
Abstract
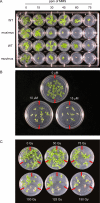
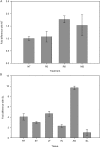
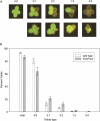
MUS81 is conserved among plants, animals, and fungi and is known to be involved in mitotic DNA damage repair and meiotic recombination. Here we present a functional characterization of the Arabidopsis thaliana homolog AtMUS81, which has a role in both mitotic and meiotic cells. The AtMUS81 transcript is produced in all tissues, but is elevated greater than 9-fold in the anthers and its levels are increased in response to gamma radiation and methyl methanesulfonate treatment. An Atmus81 transfer-DNA insertion mutant shows increased sensitivity to a wide range of DNA-damaging agents, confirming its role in mitotically proliferating cells. To examine its role in meiosis, we employed a pollen tetrad-based visual assay. Data from genetic intervals on Chromosomes 1 and 3 show that Atmus81 mutants have a moderate decrease in meiotic recombination. Importantly, measurements of recombination in a pair of adjacent intervals on Chromosome 5 demonstrate that the remaining crossovers in Atmus81 are interference sensitive, and that interference levels in the Atmus81 mutant are significantly greater than those in wild type. These data are consistent with the hypothesis that AtMUS81 is involved in a secondary subset of meiotic crossovers that are interference insensitive.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures


References
-
- Szostak JW, Orr-Weaver TL, Rothstein RJ, Stahl FW. The double-strand-break repair model for recombination. Cell. 1983;33:25–35. - PubMed
-
- Zickler D, Kleckner N. The leptotene-zygotene transition of meiosis. Annu Rev Genet. 1998;32:619–697. - PubMed
-
- Sun H, Treco D, Szostak JW. Extensive 3′-overhanging, single-stranded DNA associated with the meiosis-specific double-strand breaks at the ARG4 recombination initiation site. Cell. 1991;64:1155–1161. - PubMed
-
- Keeney S, Giroux CN, Kleckner N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997;88:375–384. - PubMed
-
- Sturtevant AH. The behavior of chromosomes as studied through linkage. Z Induct Abstammungs-Vererbungsl. 1915;13:234–287.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

