GSK-3 inhibitors induce chromosome instability
- PMID: 17697341
- PMCID: PMC1976608
- DOI: 10.1186/1471-2121-8-34
GSK-3 inhibitors induce chromosome instability
Abstract
Background: Several mechanisms operate during mitosis to ensure accurate chromosome segregation. However, during tumour evolution these mechanisms go awry resulting in chromosome instability. While several lines of evidence suggest that mutations in adenomatous polyposis coli (APC) may promote chromosome instability, at least in colon cancer, the underlying mechanisms remain unclear. Here, we turn our attention to GSK-3 - a protein kinase, which in concert with APC, targets beta-catenin for proteolysis - and ask whether GSK-3 is required for accurate chromosome segregation.
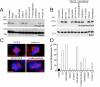
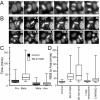
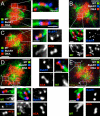
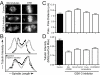
Results: To probe the role of GSK-3 in mitosis, we inhibited GSK-3 kinase activity in cells using a panel of small molecule inhibitors, including SB-415286, AR-A014418, 1-Azakenpaullone and CHIR99021. Analysis of synchronised HeLa cells shows that GSK-3 inhibitors do not prevent G1/S progression or cell division. They do, however, significantly delay mitotic exit, largely because inhibitor-treated cells have difficulty aligning all their chromosomes. Although bipolar spindles form and the majority of chromosomes biorient, one or more chromosomes often remain mono-oriented near the spindle poles. Despite a prolonged mitotic delay, anaphase frequently initiates without the last chromosome aligning, resulting in chromosome non-disjunction. To rule out the possibility of "off-target" effects, we also used RNA interference to selectively repress GSK-3beta. Cells deficient for GSK-3beta exhibit a similar chromosome alignment defect, with chromosomes clustered near the spindle poles. GSK-3beta repression also results in cells accumulating micronuclei, a hallmark of chromosome missegregation.
Conclusion: Thus, not only do our observations indicate a role for GSK-3 in accurate chromosome segregation, but they also raise the possibility that, if used as therapeutic agents, GSK-3 inhibitors may induce unwanted side effects by inducing chromosome instability.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

