A physical map of the bovine genome
- PMID: 17697342
- PMCID: PMC2374996
- DOI: 10.1186/gb-2007-8-8-r165
A physical map of the bovine genome
Abstract
Background: Cattle are important agriculturally and relevant as a model organism. Previously described genetic and radiation hybrid (RH) maps of the bovine genome have been used to identify genomic regions and genes affecting specific traits. Application of these maps to identify influential genetic polymorphisms will be enhanced by integration with each other and with bacterial artificial chromosome (BAC) libraries. The BAC libraries and clone maps are essential for the hybrid clone-by-clone/whole-genome shotgun sequencing approach taken by the bovine genome sequencing project.
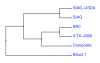
Results: A bovine BAC map was constructed with HindIII restriction digest fragments of 290,797 BAC clones from animals of three different breeds. Comparative mapping of 422,522 BAC end sequences assisted with BAC map ordering and assembly. Genotypes and pedigree from two genetic maps and marker scores from three whole-genome RH panels were consolidated on a 17,254-marker composite map. Sequence similarity allowed integrating the BAC and composite maps with the bovine draft assembly (Btau3.1), establishing a comprehensive resource describing the bovine genome. Agreement between the marker and BAC maps and the draft assembly is high, although discrepancies exist. The composite and BAC maps are more similar than either is to the draft assembly.
Conclusion: Further refinement of the maps and greater integration into the genome assembly process may contribute to a high quality assembly. The maps provide resources to associate phenotypic variation with underlying genomic variation, and are crucial resources for understanding the biology underpinning this important ruminant species so closely associated with humans.
Figures

References
-
- Diamond J. Guns, Germs and Steel: the Fates of Human Societies. New York, NY, USA: WW Norton & Company; 1997.
-
- Willham RL. From husbandry to science: A highly significant facet of our livestock heritage. J Anim Sci. 1986;62:1742–1758.
-
- Lynch M, Walsh B. Genetics and Analysis of Quantitative Traits. Sunderland, MA, USA: Sinauer Associates, Inc; 1998.
-
- Mason IL. World Dictionary of Livestock Breeds. 3. Wallingford, UK: CAB International; 1998.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

