Selection of reference genes for gene expression studies in pig tissues using SYBR green qPCR
- PMID: 17697375
- PMCID: PMC2000887
- DOI: 10.1186/1471-2199-8-67
Selection of reference genes for gene expression studies in pig tissues using SYBR green qPCR
Abstract
Background: Real-time quantitative PCR (qPCR) is a method for rapid and reliable quantification of mRNA transcription. Internal standards such as reference genes are used to normalise mRNA levels between different samples for an exact comparison of mRNA transcription level. Selection of high quality reference genes is of crucial importance for the interpretation of data generated by real-time qPCR.
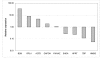
Results: In this study nine commonly used reference genes were investigated in 17 different pig tissues using real-time qPCR with SYBR green. The genes included beta-actin (ACTB), beta-2-microglobulin (B2M), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), hydroxymethylbilane synthase (HMBS), hypoxanthine phosphoribosyltransferase 1 (HPRT1), ribosomal protein L4 (RPL4), succinate dehydrogenase complex subunit A (SDHA), TATA box binding protein (TPB)and tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta polypeptide (YWHAZ). The stability of these reference genes in different pig tissues was investigated using the geNorm application. The range of expression stability in the genes analysed was (from the most stable to the least stable): ACTB/RPL4, TBP, HPRT, HMBS, YWHAZ, SDHA, B2M and GAPDH.
Conclusion: Expression stability varies greatly between genes. ACTB, RPL4, TPB and HPRT1 were found to have the highest stability across tissues. Based on both expression stability and expression level, our data suggest that ACTB and RPL4 are good reference genes for high abundant transcripts while TPB and HPRT1 are good reference genes for low abundant transcripts in expression studies across different pig tissues.
Figures

Similar articles
-
Determination of stable reference genes for RT-qPCR expression data in mechanistic pain studies on pig dorsal root ganglia and spinal cord.Res Vet Sci. 2017 Oct;114:493-501. doi: 10.1016/j.rvsc.2017.09.025. Epub 2017 Sep 28. Res Vet Sci. 2017. PMID: 28987956 Free PMC article.
-
Selection of suitable reference genes for normalization of quantitative RT-PCR (RT-qPCR) expression data across twelve tissues of riverine buffaloes (Bubalus bubalis).PLoS One. 2018 Mar 6;13(3):e0191558. doi: 10.1371/journal.pone.0191558. eCollection 2018. PLoS One. 2018. PMID: 29509770 Free PMC article.
-
A new set of reference housekeeping genes for the normalization RT-qPCR data from the intestine of piglets during weaning.PLoS One. 2018 Sep 26;13(9):e0204583. doi: 10.1371/journal.pone.0204583. eCollection 2018. PLoS One. 2018. PMID: 30256841 Free PMC article.
-
Normalization of Gene Expression by Quantitative RT-PCR in Human Cell Line: comparison of 12 Endogenous Reference Genes.Ethiop J Health Sci. 2018 Nov;28(6):741-748. doi: 10.4314/ejhs.v28i6.9. Ethiop J Health Sci. 2018. PMID: 30607091 Free PMC article.
-
Selection of reliable reference genes for qPCR studies on chondroprotective action.BMC Mol Biol. 2007 Feb 26;8:13. doi: 10.1186/1471-2199-8-13. BMC Mol Biol. 2007. PMID: 17324259 Free PMC article.
Cited by
-
Regulatory Effect of Methylation of the Porcine AQP3 Gene Promoter Region on Its Expression Level and Porcine Epidemic Diarrhea Virus Resistance.Genes (Basel). 2020 Oct 6;11(10):1167. doi: 10.3390/genes11101167. Genes (Basel). 2020. PMID: 33036186 Free PMC article.
-
Potential of Oral Nanoparticles Containing Cytokines as Intestinal Mucosal Immunostimulants in Pigs: A Pilot Study.Animals (Basel). 2022 Apr 21;12(9):1075. doi: 10.3390/ani12091075. Animals (Basel). 2022. PMID: 35565502 Free PMC article.
-
Immunobiotic Lactobacillus jensenii as immune-health promoting factor to improve growth performance and productivity in post-weaning pigs.BMC Immunol. 2014 Jun 19;15:24. doi: 10.1186/1471-2172-15-24. BMC Immunol. 2014. PMID: 24943108 Free PMC article.
-
In vitro and ex vivo analyses of co-infections with swine influenza and porcine reproductive and respiratory syndrome viruses.Vet Microbiol. 2014 Feb 21;169(1-2):18-32. doi: 10.1016/j.vetmic.2013.11.037. Epub 2013 Dec 14. Vet Microbiol. 2014. PMID: 24418046 Free PMC article.
-
Selection of Reference Genes for Gene Expression Studies in Porcine Whole Blood and Peripheral Blood Mononuclear Cells under Polyinosinic:Polycytidylic Acid Stimulation.Asian-Australas J Anim Sci. 2014 Apr;27(4):471-8. doi: 10.5713/ajas.2013.13471. Asian-Australas J Anim Sci. 2014. PMID: 25049976 Free PMC article.
References
-
- de Kok JB, Roelofs RW, Giesendorf BA, Pennings JL, Waas ET, Feuth T, Swinkels DW, Span PN. Normalization of gene expression measurements in tumor tissues: comparison of 13 endogenous control genes. Lab Invest. 2005;85:154–159. - PubMed
-
- Erkens T, Van Poucke M, Vandesompele J, Goossens K, Van Zeveren A, Peelman LJ. Development of a new set of reference genes for normalization of real-time RT-PCR data of porcine backfat and longissimus dorsi muscle, and evaluation with PPARGC1A. BMC Biotechnol. 2006;6:41. doi: 10.1186/1472-6750-6-41. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

