One-step selection of Vaccinia virus-binding DNA aptamers by MonoLEX
- PMID: 17697378
- PMCID: PMC1994675
- DOI: 10.1186/1472-6750-7-48
One-step selection of Vaccinia virus-binding DNA aptamers by MonoLEX
Abstract
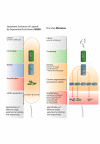
Background: As a new class of therapeutic and diagnostic reagents, more than fifteen years ago RNA and DNA aptamers were identified as binding molecules to numerous small compounds, proteins and rarely even to complete pathogen particles. Most aptamers were isolated from complex libraries of synthetic nucleic acids by a process termed SELEX based on several selection and amplification steps. Here we report the application of a new one-step selection method (MonoLEX) to acquire high-affinity DNA aptamers binding Vaccinia virus used as a model organism for complex target structures.
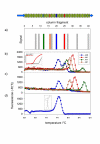
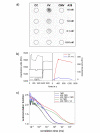
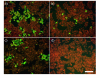
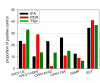
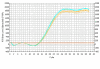
Results: The selection against complete Vaccinia virus particles resulted in a 64-base DNA aptamer specifically binding to orthopoxviruses as validated by dot blot analysis, Surface Plasmon Resonance, Fluorescence Correlation Spectroscopy and real-time PCR, following an aptamer blotting assay. The same oligonucleotide showed the ability to inhibit in vitro infection of Vaccinia virus and other orthopoxviruses in a concentration-dependent manner.
Conclusion: The MonoLEX method is a straightforward procedure as demonstrated here for the identification of a high-affinity DNA aptamer binding Vaccinia virus. MonoLEX comprises a single affinity chromatography step, followed by subsequent physical segmentation of the affinity resin and a single final PCR amplification step of bound aptamers. Therefore, this procedure improves the selection of high affinity aptamers by reducing the competition between aptamers of different affinities during the PCR step, indicating an advantage for the single-round MonoLEX method.
Figures
References
-
- Jahrling PB, Fritz EA, Hensley LE. Countermeasures to the bioterrorist threat of smallpox. Curr Mol Med. 2005;5:817–826. - PubMed
-
- Yang G, Pevear DC, Davies MH, Collett MS, Bailey T, Rippen S, Barone L, Burns C, Rhodes G, Tohan S, Huggins JW, Baker RO, Buller RL, Touchette E, Waller K, Schriewer J, Neyts J, DeClercq E, Jones K, Hruby D, Jordan R. An orally bioavailable antipoxvirus compound (ST-246) inhibits extracellular virus formation and protects mice from lethal orthopoxvirus Challenge. J Virol. 2005;79:13139–13149. - PMC - PubMed
-
- Zhang Z, Blank M, Schluesener HJ. Nucleic acid aptamers in human viral disease. Arch Immunol Ther Exp (Warsz ) 2004;52:307–315. - PubMed
-
- Breaker RR. Natural and engineered nucleic acids as tools to explore biology. Nature. 2004;432:838–845. - PubMed
-
- Hesselberth JR, Miller D, Robertus J, Ellington AD. In vitro selection of RNA molecules that inhibit the activity of ricin A-chain. J Biol Chem. 2000;275:4937–4942. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

