BCoR-L1 variation and breast cancer
- PMID: 17697391
- PMCID: PMC2206730
- DOI: 10.1186/bcr1759
BCoR-L1 variation and breast cancer
Erratum in
- Breast Cancer Res. 2008;10(5):406
Abstract
Introduction: BRCA1 is involved in numerous essential processes in the cell, and the effects of BRCA1 dysfunction in breast cancer carcinogenesis are well described. Many of the breast cancer susceptibility genes such as BRCA2, p53, ATM, CHEK2, and BRIP1 encode proteins that interact with BRCA1. BCL6 corepressor-like 1 (BCoR-L1) is a newly described BRCA1-interacting protein that displays high homology to several proteins known to be involved in the fundamental processes of DNA damage repair and transcription regulation. BCoR-L1 has been shown to play a role in transcription corepression, and expression of the X-linked BCoR-L1 gene has been reported to be dysregulated in breast cancer subjects. BCoR-L1 is located on the X chromosome and is subject to X inactivation.
Methods: We performed mutation analysis of 38 BRCA1/2 mutation-negative breast cancer families with male breast cancer, prostate cancer, and/or haplotype sharing around BCoR-L1 to determine whether there is a role for BCoR-L1 as a high-risk breast cancer predisposition gene. In addition, we conducted quantitative real-time PCR (qRT-PCR) on lymphoblastoid cell lines (LCLs) from the index cases from these families and a number of cancer cell lines to assess the role of BCoR-L1 dysregulation in cancer and cancer families.
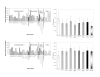
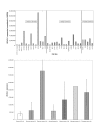
Results: Very little variation was detected in the coding region, and qRT-PCR analysis revealed that BCoR-L1 expression is highly variable in cancer-free subjects, high-risk breast cancer patients, and cancer cell lines. We also report the investigation of a new expression control, DIDO1 (death inducer-obliterator 1), that is superior to GAPDH (glyceraldehyde-3-phosphate dehydrogenase) and UBC (ubiquitin C) for analysis of expression in LCLs.
Conclusion: Our results suggest that BCoR-L1 expression does not play a large role in predisposition to familial breast cancer.
Figures
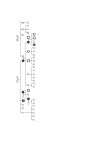
 = breast cancer-positive; c.516T>C and c.3608-156C>T-positive.
= breast cancer-positive; c.516T>C and c.3608-156C>T-positive.  = breast cancer-negative; c.516T>C and c.3608-156C>T-positive. □ = breast cancer-negative; c.516T>C and c.3608-156C>T-negative. Circle = female, square = male; subjects marked by small shapes were not available for genotyping. BCoR-L1, BCL6 corepressor-like 1.
= breast cancer-negative; c.516T>C and c.3608-156C>T-positive. □ = breast cancer-negative; c.516T>C and c.3608-156C>T-negative. Circle = female, square = male; subjects marked by small shapes were not available for genotyping. BCoR-L1, BCL6 corepressor-like 1.
Similar articles
-
Mutational analysis of the BRCA1-interacting genes ZNF350/ZBRK1 and BRIP1/BACH1 among BRCA1 and BRCA2-negative probands from breast-ovarian cancer families and among early-onset breast cancer cases and reference individuals.Hum Mutat. 2003 Aug;22(2):121-8. doi: 10.1002/humu.10238. Hum Mutat. 2003. PMID: 12872252
-
Skewed X chromosome inactivation and breast and ovarian cancer status: evidence for X-linked modifiers of BRCA1.J Natl Cancer Inst. 2008 Nov 5;100(21):1519-29. doi: 10.1093/jnci/djn345. Epub 2008 Oct 28. J Natl Cancer Inst. 2008. PMID: 18957670
-
BRCA1 and BRCA2 have a limited role in familial prostate cancer.Cancer Res. 2000 Mar 1;60(5):1371-5. Cancer Res. 2000. PMID: 10728701
-
BRCA1 and BRCA2: the genetic testing and the current management options for mutation carriers.Crit Rev Oncol Hematol. 2006 Jan;57(1):1-23. doi: 10.1016/j.critrevonc.2005.05.003. Epub 2005 Dec 6. Crit Rev Oncol Hematol. 2006. PMID: 16337408 Review.
-
Breast cancer susceptibility testing: past, present and future.Expert Rev Anticancer Ther. 2006 Aug;6(8):1205-14. doi: 10.1586/14737140.6.8.1205. Expert Rev Anticancer Ther. 2006. PMID: 16925486 Review.
Cited by
-
BCORL1 is an independent prognostic marker and contributes to cell migration and invasion in human hepatocellular carcinoma.BMC Cancer. 2016 Feb 15;16:103. doi: 10.1186/s12885-016-2154-z. BMC Cancer. 2016. Retraction in: BMC Cancer. 2023 Jun 13;23(1):542. doi: 10.1186/s12885-023-11051-6. PMID: 26879601 Free PMC article. Retracted.
References
-
- Ford D, Easton DF, Stratton M, Narod S, Goldgar D, Devilee P, Bishop DT, Weber B, Lenoir G, Chang-Claude J, et al. Genetic heterogeneity and penetrance analysis of the BRCA1 and BRCA2 genes in breast cancer families. The Breast Cancer Linkage Consortium. Am J Hum Genet. 1998;62:676–689. doi: 10.1086/301749. - DOI - PMC - PubMed
-
- Sidransky D, Tokino T, Helzlsouer K, Zehnbauer B, Rausch G, Shelton B, Prestigiacomo L, Vogelstein B, Davidson N. Inherited p53 gene mutations in breast cancer. Cancer Res. 1992;52:2984–2986. - PubMed
-
- Chenevix-Trench G, Spurdle AB, Gatei M, Kelly H, Marsh A, Chen X, Donn K, Cummings M, Nyholt D, Jenkins MA, et al. Dominant negative ATM mutations in breast cancer families. J Natl Cancer Inst. 2002;94:205–215. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

