NikD, an unusual amino acid oxidase essential for nikkomycin biosynthesis: structures of closed and open forms at 1.15 and 1.90 A resolution
- PMID: 17697998
- PMCID: PMC2764521
- DOI: 10.1016/j.str.2007.06.010
NikD, an unusual amino acid oxidase essential for nikkomycin biosynthesis: structures of closed and open forms at 1.15 and 1.90 A resolution
Abstract
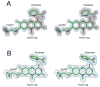
NikD is an unusual amino-acid-oxidizing enzyme that contains covalently bound FAD, catalyzes a 4-electron oxidation of piperideine-2-carboxylic acid to picolinate, and plays a critical role in the biosynthesis of nikkomycin antibiotics. Crystal structures of closed and open forms of nikD, a two-domain enzyme, have been determined to resolutions of 1.15 and 1.9 A, respectively. The two forms differ by an 11 degrees rotation of the catalytic domain with respect to the FAD-binding domain. The active site is inaccessible to solvent in the closed form; an endogenous ligand, believed to be picolinate, is bound close to and parallel with the flavin ring, an orientation compatible with redox catalysis. The active site is solvent accessible in the open form, but the picolinate ligand is approximately perpendicular to the flavin ring and a tryptophan is stacked above the flavin ring. NikD also contains a mobile cation binding loop.
Figures



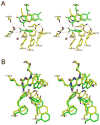


References
-
- Barriere JC, Berthaud N, Beyer D, Dutka-Malen S, Paris JM, Desnottes JF. Recent developments in streptogramin research. Curr Pharm Des. 1998;4:155–180. - PubMed
-
- Bormann C, Mattern S, Schrempf H, Fiedler HP, Zahner H. Isolation of streptomyces tendae mutants with an altered nikkomycin spectrum. J Antibiot. 1989;42:913–918. - PubMed
-
- Bruckner RC, Zhao G, Venci D, Jorns MS. Nikkomycin biosynthesis: Formation of a 4-electron oxidation product during turnover of nikD with its physiological substrate. Biochemistry. 2004;43:9160–9167. - PubMed
-
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & nmr system: A new software suite for macromolecular structure determination. Acta Cryst. 1998;D 54:905–921. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

