Presenilins regulate alphabeta T cell development by modulating TCR signaling
- PMID: 17698590
- PMCID: PMC2118698
- DOI: 10.1084/jem.20070550
Presenilins regulate alphabeta T cell development by modulating TCR signaling
Abstract
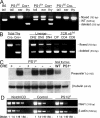
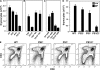
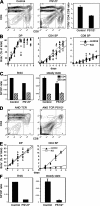
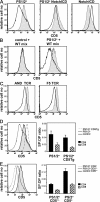
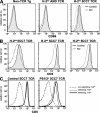
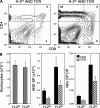
TCRalphabeta signaling is crucial for the maturation of CD4 and CD8 T cells, but the role of the Notch signaling pathway in this process is poorly understood. Genes encoding Presenilin (PS) 1/2 were deleted to prevent activation of the multiple Notch receptors expressed by developing thymocytes. PS1/2 knockout thymocyte precursors inefficiently generate CD4 T cells, a phenotype that is most pronounced when thymocytes bear a single major histocompatibility complex (MHC) class II-restricted T cell receptor (TCR). Diminished T cell production correlated with evidence of impaired TCR signaling, and could be rescued by manipulations that enhance MHC recognition. Although Notch appears to directly regulate binary fate decisions in many systems, these findings suggest a model in which PS-dependent Notch signaling influences positive selection and the development of alphabeta T cells by modifying TCR signal transduction.
Figures

Similar articles
-
Signal transduction via CD4, CD8, and CD28 in mature and immature thymocytes. Implications for thymic selection.J Immunol. 1991 Mar 1;146(5):1428-36. J Immunol. 1991. PMID: 1847160
-
CD4 and CD8 are positive regulators of T cell receptor signal transduction in early T cell differentiation.J Immunol. 1991 Mar 15;146(6):1759-65. J Immunol. 1991. PMID: 1706380
-
Critical relationship between TCR signaling potential and TCR affinity during thymocyte selection.J Immunol. 2000 Sep 15;165(6):3080-7. doi: 10.4049/jimmunol.165.6.3080. J Immunol. 2000. PMID: 10975819
-
TCR and Notch signaling in CD4 and CD8 T-cell development.Immunol Rev. 2006 Feb;209:274-83. doi: 10.1111/j.0105-2896.2006.00358.x. Immunol Rev. 2006. PMID: 16448548 Review.
-
CD4 function in thymocyte differentiation and T cell activation.Philos Trans R Soc Lond B Biol Sci. 1993 Oct 29;342(1299):25-34. doi: 10.1098/rstb.1993.0131. Philos Trans R Soc Lond B Biol Sci. 1993. PMID: 7904343 Review.
Cited by
-
CD4 CTL: living up to the challenge.Semin Immunol. 2013 Nov 15;25(4):273-81. doi: 10.1016/j.smim.2013.10.022. Epub 2013 Nov 15. Semin Immunol. 2013. PMID: 24246226 Free PMC article. Review.
-
Notch Partners in the Long Journey of T-ALL Pathogenesis.Int J Mol Sci. 2023 Jan 10;24(2):1383. doi: 10.3390/ijms24021383. Int J Mol Sci. 2023. PMID: 36674902 Free PMC article. Review.
-
Alternative pathway for the development of Vα14+ NKT cells directly from CD4-CD8- thymocytes that bypasses the CD4+CD8+ stage.Nat Immunol. 2017 Mar;18(3):274-282. doi: 10.1038/ni.3668. Epub 2017 Jan 30. Nat Immunol. 2017. PMID: 28135253
-
Notch and presenilin regulate cellular expansion and cytokine secretion but cannot instruct Th1/Th2 fate acquisition.PLoS One. 2008 Jul 30;3(7):e2823. doi: 10.1371/journal.pone.0002823. PLoS One. 2008. PMID: 18665263 Free PMC article.
-
Defective signal transduction in B lymphocytes lacking presenilin proteins.Proc Natl Acad Sci U S A. 2008 Jan 22;105(3):979-84. doi: 10.1073/pnas.0707755105. Epub 2008 Jan 14. Proc Natl Acad Sci U S A. 2008. PMID: 18195359 Free PMC article.
References
-
- Robey, E., and B.J. Fowlkes. 1994. Selective events in T cell development. Annu. Rev. Immunol. 12:675–705. - PubMed
-
- Germain, R.N. 2002. T-cell development and the CD4-CD8 lineage decision. Nat. Rev. Immunol. 2:309–322. - PubMed
-
- Bosselut, R. 2004. CD4/CD8-lineage differentiation in the thymus: from nuclear effectors to membrane signals. Nat. Rev. Immunol. 4:529–540. - PubMed
-
- Ciofani, M., T.M. Schmitt, A. Ciofani, A.M. Michie, N. Cuburu, A. Aublin, J.L. Maryanski, and J.C. Zuniga-Pflucker. 2004. Obligatory role for cooperative signaling by pre-TCR and notch during thymocyte differentiation. J. Immunol. 172:5230–5239. - PubMed
-
- Wolfer, A., A. Wilson, M. Nemir, H.R. MacDonald, and F. Radtke. 2002. Inactivation of Notch1 impairs VDJβ rearrangement and allows pre-TCR-independent survival of early αβ lineage thymocytes. Immunity. 16:869–879. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

