An in vitro nuclear disassembly system reveals a role for the RanGTPase system and microtubule-dependent steps in nuclear envelope breakdown
- PMID: 17698605
- PMCID: PMC2064467
- DOI: 10.1083/jcb.200703002
An in vitro nuclear disassembly system reveals a role for the RanGTPase system and microtubule-dependent steps in nuclear envelope breakdown
Abstract
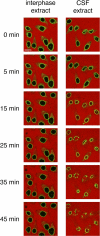
During prophase, vertebrate cells disassemble their nuclear envelope (NE) in the process of NE breakdown (NEBD). We have established an in vitro assay that uses mitotic Xenopus laevis egg extracts and semipermeabilized somatic cells bearing a green fluorescent protein-tagged NE marker to study the molecular requirements underlying the dynamic changes of the NE during NEBD by live microscopy. We applied our in vitro system to analyze the role of the Ran guanosine triphosphatase (GTPase) system in NEBD. Our study shows that high levels of RanGTP affect the dynamics of late steps of NEBD in vitro. Also, inhibition of RanGTP production by RanT24N blocks the dynamic rupture of nuclei, suggesting that the local generation of RanGTP around chromatin may serve as a spatial cue in NEBD. Furthermore, the microtubule-depolymerizing drug nocodazole interferes with late steps of nuclear disassembly in vitro. High resolution live cell imaging reveals that microtubules are involved in the completion of NEBD in vivo by facilitating the efficient removal of membranes from chromatin.
Figures



Similar articles
-
Analysis of a RanGTP-regulated gradient in mitotic somatic cells.Nature. 2006 Mar 30;440(7084):697-701. doi: 10.1038/nature04589. Nature. 2006. PMID: 16572176
-
Characterization of Ran-driven cargo transport and the RanGTPase system by kinetic measurements and computer simulation.EMBO J. 2003 Mar 3;22(5):1088-100. doi: 10.1093/emboj/cdg113. EMBO J. 2003. PMID: 12606574 Free PMC article.
-
An in vitro system to study nuclear envelope breakdown.Methods Cell Biol. 2014;122:255-76. doi: 10.1016/B978-0-12-417160-2.00012-6. Methods Cell Biol. 2014. PMID: 24857734
-
Spatial and temporal control of nuclear envelope assembly by Ran GTPase.Symp Soc Exp Biol. 2004;(56):193-204. Symp Soc Exp Biol. 2004. PMID: 15565882 Review.
-
Composition of the plant nuclear envelope: theme and variations.J Exp Bot. 2007;58(1):27-34. doi: 10.1093/jxb/erl009. Epub 2006 Jun 27. J Exp Bot. 2007. PMID: 16804054 Review.
Cited by
-
Mitotic lamin disassembly is triggered by lipid-mediated signaling.J Cell Biol. 2012 Sep 17;198(6):981-90. doi: 10.1083/jcb.201205103. J Cell Biol. 2012. PMID: 22986494 Free PMC article.
-
Border control at the nucleus: biogenesis and organization of the nuclear membrane and pore complexes.Dev Cell. 2009 Nov;17(5):606-16. doi: 10.1016/j.devcel.2009.10.007. Dev Cell. 2009. PMID: 19922866 Free PMC article. Review.
-
Farnesol, a fungal quorum-sensing molecule triggers apoptosis in human oral squamous carcinoma cells.Neoplasia. 2008 Sep;10(9):954-63. doi: 10.1593/neo.08444. Neoplasia. 2008. PMID: 18714396 Free PMC article.
-
Nuclear shape, mechanics, and mechanotransduction.Circ Res. 2008 Jun 6;102(11):1307-18. doi: 10.1161/CIRCRESAHA.108.173989. Circ Res. 2008. PMID: 18535268 Free PMC article. Review.
-
The biochemistry of mitosis.Cold Spring Harb Perspect Biol. 2015 Feb 6;7(3):a015776. doi: 10.1101/cshperspect.a015776. Cold Spring Harb Perspect Biol. 2015. PMID: 25663668 Free PMC article. Review.
References
-
- Beaudouin, J., D. Gerlich, N. Daigle, R. Eils, and J. Ellenberg. 2002. Nuclear envelope breakdown proceeds by microtubule-induced tearing of the lamina. Cell. 108:83–96. - PubMed
-
- Busson, S., D. Dujardin, A. Moreau, J. Dompierre, and J.R. De Mey. 1998. Dynein and dynactin are localized to astral microtubules and at cortical sites in mitotic epithelial cells. Curr. Biol. 8:541–544. - PubMed
-
- Carazo-Salas, R.E., G. Guarguaglini, O.J. Gruss, A. Segref, E. Karsenti, and I.W. Mattaj. 1999. Generation of GTP-bound Ran by RCC1 is required for chromatin-induced mitotic spindle formation. Nature. 400:178–181. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

