Instrumentation and metrology for single RNA counting in biological complexes or nanoparticles by a single-molecule dual-view system
- PMID: 17698643
- PMCID: PMC1986819
- DOI: 10.1261/rna.587607
Instrumentation and metrology for single RNA counting in biological complexes or nanoparticles by a single-molecule dual-view system
Abstract
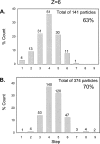
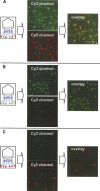
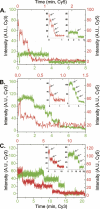
Limited by the spatial resolution of optical microscopy, direct detection or counting of single components in biological complexes or nanoparticles is challenging, especially for RNA, which is conformationally versatile and structurally flexible. We report here the assembly of a customized single-molecule dual-viewing total internal reflection fluorescence imaging system for direct counting of RNA building blocks. The RNA molecules were labeled with a single fluorophore by in vitro transcription in the presence of a fluorescent AMP. Precise calculation of identical or mixed pRNA building blocks of one, two, three, or six copies within the bacteriophage phi29 DNA packaging motor or other complexes was demonstrated by applying a photobleaching assay and evaluated by binomial distribution. The dual-viewing system for excitation and recording at different wavelengths simultaneously will enable the differentiation of different complexes with different labels or relative motion of each labeled component in motion machines.
Figures




References
-
- Ambrose, W.P., Goodwin, P.M., Nolan, J.P. Single-molecule detection with total internal reflection excitation: Comparing signal-to-background and total signals in different geometries. Cytometry. 1999;36:224–231. - PubMed
-
- Block, S.M., Goldstein, L.S., Schnapp, B.J. Bead movement by single kinesin molecules studied with optical tweezers. Nature. 1990;348:348–352. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
