Structural basis for the acyl chain selectivity and mechanism of UDP-N-acetylglucosamine acyltransferase
- PMID: 17698807
- PMCID: PMC1959417
- DOI: 10.1073/pnas.0705833104
Structural basis for the acyl chain selectivity and mechanism of UDP-N-acetylglucosamine acyltransferase
Abstract
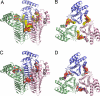
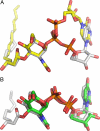
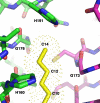
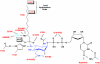
UDP-N-acetylglucosamine (UDP-GlcNAc) acyltransferase (LpxA) catalyzes the first step of lipid A biosynthesis, the reversible transfer of the R-3-hydroxyacyl chain from R-3-hydroxyacyl acyl carrier protein to the glucosamine 3-OH group of UDP-GlcNAc. Escherichia coli LpxA is highly selective for R-3-hydroxymyristate. The crystal structure of the E. coli LpxA homotrimer, determined previously in the absence of lipid substrates or products, revealed that LpxA contains an unusual, left-handed parallel beta-helix fold. We have now solved the crystal structures of E. coli LpxA with the bound product UDP-3-O-(R-3-hydroxymyristoyl)-GlcNAc at a resolution of 1.74 A and with bound UDP-3-O-(R-3-hydroxydecanoyl)-GlcNAc at 1.85 A. The structures of these complexes are consistent with the catalytic mechanism deduced by mutagenesis and with a recent 3.0-A structure of LpxA with bound UDP-GlcNAc. Our structures show how LpxA selects for 14-carbon R-3-hydroxyacyl chains and reveal two modes of UDP binding.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

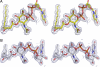

Similar articles
-
Structure of UDP-N-acetylglucosamine acyltransferase with a bound antibacterial pentadecapeptide.Proc Natl Acad Sci U S A. 2006 Jul 18;103(29):10877-82. doi: 10.1073/pnas.0604465103. Epub 2006 Jul 11. Proc Natl Acad Sci U S A. 2006. PMID: 16835299 Free PMC article.
-
Structures of Pseudomonas aeruginosa LpxA Reveal the Basis for Its Substrate Selectivity.Biochemistry. 2015 Sep 29;54(38):5937-48. doi: 10.1021/acs.biochem.5b00720. Epub 2015 Sep 18. Biochemistry. 2015. PMID: 26352800
-
Crystal structure and activity of Francisella novicida UDP-N-acetylglucosamine acyltransferase.Biochem Biophys Res Commun. 2016 Sep 23;478(3):1223-9. doi: 10.1016/j.bbrc.2016.08.098. Epub 2016 Aug 19. Biochem Biophys Res Commun. 2016. PMID: 27545601 Free PMC article.
-
Lipid a biosynthesis of multidrug-resistant pathogens - a novel drug target.Curr Pharm Des. 2013;19(36):6534-50. doi: 10.2174/13816128113199990494. Curr Pharm Des. 2013. PMID: 23829374 Review.
-
The enzymes of neutral lipid synthesis.J Biol Chem. 2001 Nov 2;276(44):40369-72. doi: 10.1074/jbc.R100050200. Epub 2001 Sep 5. J Biol Chem. 2001. PMID: 11544264 Review. No abstract available.
Cited by
-
Evolution of the Kdo2-lipid A biosynthesis in bacteria.BMC Evol Biol. 2010 Nov 24;10:362. doi: 10.1186/1471-2148-10-362. BMC Evol Biol. 2010. PMID: 21106097 Free PMC article.
-
Crystal structure and functional implications of the tandem-type universal stress protein UspE from Escherichia coli.BMC Struct Biol. 2016 Feb 11;16:3. doi: 10.1186/s12900-016-0053-9. BMC Struct Biol. 2016. PMID: 26865045 Free PMC article.
-
Current Progress in the Structural and Biochemical Characterization of Proteins Involved in the Assembly of Lipopolysaccharide.Int J Microbiol. 2018 Nov 25;2018:5319146. doi: 10.1155/2018/5319146. eCollection 2018. Int J Microbiol. 2018. PMID: 30595696 Free PMC article. Review.
-
Antibacterial activities of anthraquinones: structure-activity relationships and action mechanisms.RSC Med Chem. 2023 Jul 10;14(8):1446-1471. doi: 10.1039/d3md00116d. eCollection 2023 Aug 16. RSC Med Chem. 2023. PMID: 37593578 Free PMC article. Review.
-
Ecogenomics reveals viral communities across the Challenger Deep oceanic trench.Commun Biol. 2022 Oct 4;5(1):1055. doi: 10.1038/s42003-022-04027-y. Commun Biol. 2022. PMID: 36192584 Free PMC article.
References
-
- Rietschel ET, Kirikae T, Schade FU, Mamat U, Schmidt G, Loppnow H, Ulmer AJ, Zähringer U, Seydel U, Di Padova F, et al. FASEB J. 1994;8:217–225. - PubMed
-
- Raetz CRH, Garrett TA, Reynolds CM, Shaw WA, Moore JD, Smith DC, Jr, Ribeiro AA, Murphy RC, Ulevitch RJ, Fearns C, et al. J Lipid Res. 2006;47:1097–1111. - PubMed
-
- Anderson MS, Bulawa CE, Raetz CRH. J Biol Chem. 1985;260:15536–15541. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

