Regulation of the formin for3p by cdc42p and bud6p
- PMID: 17699595
- PMCID: PMC1995706
- DOI: 10.1091/mbc.e07-02-0094
Regulation of the formin for3p by cdc42p and bud6p
Abstract
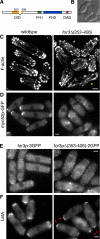
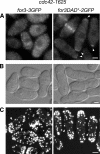
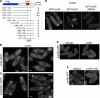
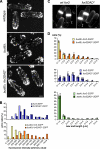
Formins are conserved actin nucleators responsible for the assembly of diverse actin structures. Many formins are controlled through an autoinhibitory mechanism involving the interaction of a C-terminal DAD sequence with an N-terminal DID sequence. Here, we show that the fission yeast formin for3p, which mediates actin cable assembly and polarized cell growth, is regulated by a similar autoinhibitory mechanism in vivo. Multiple sites govern for3p localization to cell tips. The localization and activity of for3p are inhibited by an intramolecular interaction of divergent DAD and DID-like sequences. A for3p DAD mutant expressed at endogenous levels produces more robust actin cables, which appear to have normal organization and dynamics. We identify cdc42p as the primary Rho GTPase involved in actin cable assembly and for3p regulation. Both cdc42p, which binds at the N terminus of for3p, and bud6p, which binds near the C-terminal DAD-like sequence, are needed for for3p localization and full activity, but a mutation in the for3p DAD restores for3p localization and other phenotypes of cdc42 and bud6 mutants. In particular, the for3p DAD mutation suppresses the bipolar growth (NETO) defect of bud6Delta cells. These findings suggest that cdc42p and bud6p activate for3p by relieving autoinhibition.
Figures
Similar articles
-
Dynamics of the formin for3p in actin cable assembly.Curr Biol. 2006 Jun 20;16(12):1161-70. doi: 10.1016/j.cub.2006.04.040. Curr Biol. 2006. PMID: 16782006
-
Model of For3p-mediated actin cable assembly in fission yeast.PLoS One. 2008;3(12):e4078. doi: 10.1371/journal.pone.0004078. Epub 2008 Dec 31. PLoS One. 2008. PMID: 19116660 Free PMC article.
-
Roles of the fission yeast formin for3p in cell polarity, actin cable formation and symmetric cell division.Curr Biol. 2001 Oct 30;11(21):1656-65. doi: 10.1016/s0960-9822(01)00525-5. Curr Biol. 2001. PMID: 11696322
-
Formin proteins: a domain-based approach.Trends Biochem Sci. 2005 Jun;30(6):342-53. doi: 10.1016/j.tibs.2005.04.014. Trends Biochem Sci. 2005. PMID: 15950879 Review.
-
FHOD proteins in actin dynamics--a formin' class of its own.Small GTPases. 2014;5(2):11. doi: 10.4161/21541248.2014.973765. Small GTPases. 2014. PMID: 25483300 Free PMC article. Review.
Cited by
-
Mathematical model for growth regulation of fission yeast Schizosaccharomyces pombe.PLoS One. 2012;7(11):e49675. doi: 10.1371/journal.pone.0049675. Epub 2012 Nov 27. PLoS One. 2012. PMID: 23209589 Free PMC article.
-
Oscillatory dynamics of Cdc42 GTPase in the control of polarized growth.Science. 2012 Jul 13;337(6091):239-43. doi: 10.1126/science.1218377. Epub 2012 May 17. Science. 2012. PMID: 22604726 Free PMC article.
-
Fission yeast sec3 bridges the exocyst complex to the actin cytoskeleton.Traffic. 2012 Nov;13(11):1481-95. doi: 10.1111/j.1600-0854.2012.01408.x. Epub 2012 Sep 7. Traffic. 2012. PMID: 22891673 Free PMC article.
-
Regulation of the formin Bnr1 by septins anda MARK/Par1-family septin-associated kinase.Mol Biol Cell. 2012 Oct;23(20):4041-53. doi: 10.1091/mbc.E12-05-0395. Epub 2012 Aug 23. Mol Biol Cell. 2012. PMID: 22918953 Free PMC article.
-
The functionally distinct fission yeast formins have specific actin-assembly properties.Mol Biol Cell. 2011 Oct;22(20):3826-39. doi: 10.1091/mbc.E11-06-0492. Epub 2011 Aug 24. Mol Biol Cell. 2011. PMID: 21865598 Free PMC article.
References
-
- Alberts A. S. Identification of a carboxyl-terminal diaphanous-related formin homology protein autoregulatory domain. J. Biol. Chem. 2001;276:2824–2830. - PubMed
-
- Cadwell R. C., Joyce G. F. Randomization of genes by PCR mutagenesis. PCR Methods Appl. 1992;2:28–33. - PubMed
-
- Eisenmann K. M., Harris E. S., Kitchen S. M., Holman H. A., Higgs H. N., Alberts A. S. Dia-interacting protein modulates formin-mediated actin assembly at the cell cortex. Curr. Biol. 2007;17:579–591. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

