On the importance of a funneled energy landscape for the assembly and regulation of multidomain Src tyrosine kinases
- PMID: 17699616
- PMCID: PMC1959435
- DOI: 10.1073/pnas.0704041104
On the importance of a funneled energy landscape for the assembly and regulation of multidomain Src tyrosine kinases
Abstract
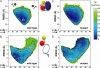
Regulation of signaling pathways in the cell often involves multidomain allosteric enzymes that are able to adopt alternate active or inactive conformations in response to specific stimuli. It is therefore of great interest to elucidate the energetic and structural determinants that govern the conformational plasticity of these proteins. In this study, free-energy computations have been used to address this fundamental question, focusing on one important family of signaling enzymes, the Src tyrosine kinases. Inactivation of these enzymes depends on the formation of an assembly comprising a tandem of SH3 and SH2 modules alongside a catalytic domain. Activation results from the release of the SH3 and SH2 domains, which are then believed to be structurally uncoupled by virtue of a flexible peptide link. In contrast to this view, this analysis shows that inactivation depends critically on the intrinsic propensity of the SH3-SH2 tandem to adopt conformations that are conducive to the assembled inactive state, even when no interactions with the rest of the kinase are possible. This funneling of the available conformational space is encoded within the SH3-SH2 connector, which appears to have evolved to modulate the flexibility of the tandem in solution. To further substantiate this notion, we show how constitutively activating mutations in the SH3-SH2 connector shift the assembly equilibrium toward the disassembled, active state. Based on a similar analysis of several constructs of the kinase complex, we propose that assembly is characterized by the progressive optimization of the protein's conformational energy, with little or no energetic frustration.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
The Activation of c-Src Tyrosine Kinase: Conformational Transition Pathway and Free Energy Landscape.J Phys Chem B. 2017 Apr 20;121(15):3352-3363. doi: 10.1021/acs.jpcb.6b08409. Epub 2016 Oct 28. J Phys Chem B. 2017. PMID: 27715044 Free PMC article.
-
Two-state dynamics of the SH3-SH2 tandem of Abl kinase and the allosteric role of the N-cap.Proc Natl Acad Sci U S A. 2013 Sep 3;110(36):E3372-80. doi: 10.1073/pnas.1303966110. Epub 2013 Aug 19. Proc Natl Acad Sci U S A. 2013. PMID: 23959873 Free PMC article.
-
Tyrosine Kinase Activation and Conformational Flexibility: Lessons from Src-Family Tyrosine Kinases.Acc Chem Res. 2017 May 16;50(5):1193-1201. doi: 10.1021/acs.accounts.7b00012. Epub 2017 Apr 20. Acc Chem Res. 2017. PMID: 28426203 Free PMC article.
-
Src protein-tyrosine kinase structure and regulation.Biochem Biophys Res Commun. 2004 Nov 26;324(4):1155-64. doi: 10.1016/j.bbrc.2004.09.171. Biochem Biophys Res Commun. 2004. PMID: 15504335 Review.
-
Novel Roles of SH2 and SH3 Domains in Lipid Binding.Cells. 2021 May 13;10(5):1191. doi: 10.3390/cells10051191. Cells. 2021. PMID: 34068055 Free PMC article. Review.
Cited by
-
Energetics of subdomain movements and fluorescence probe solvation environment change in ATP-bound myosin.Eur Biophys J. 2008 Nov;38(1):1-12. doi: 10.1007/s00249-008-0347-3. Epub 2008 Jun 21. Eur Biophys J. 2008. PMID: 18568345
-
Activation of PKA via asymmetric allosteric coupling of structurally conserved cyclic nucleotide binding domains.Nat Commun. 2019 Sep 4;10(1):3984. doi: 10.1038/s41467-019-11930-2. Nat Commun. 2019. PMID: 31484930 Free PMC article.
-
A coupled equilibrium shift mechanism in calmodulin-mediated signal transduction.Structure. 2008 May;16(5):736-46. doi: 10.1016/j.str.2008.02.017. Structure. 2008. PMID: 18462678 Free PMC article.
-
Flexibility and charge asymmetry in the activation loop of Src tyrosine kinases.Proteins. 2009 Feb 1;74(2):378-89. doi: 10.1002/prot.22153. Proteins. 2009. PMID: 18623061 Free PMC article.
-
Structural basis for the recognition of c-Src by its inactivator Csk.Cell. 2008 Jul 11;134(1):124-34. doi: 10.1016/j.cell.2008.05.051. Cell. 2008. PMID: 18614016 Free PMC article.
References
-
- Pawson T, Nash P. Science. 2003;300:445–452. - PubMed
-
- Frame MC. Biochim Biophys Acta. 2002;1602:114–130. - PubMed
-
- Harrison SC. Cell. 2003;112:737–740. - PubMed
-
- Cesareni G, Gimona M, Sudol M, Yaffe M. Weinhein, Germany: Wiley–VCH; 2005.
-
- Young MA, Gonfloni S, Superti-Furga G, Roux B, Kuriyan J. Cell. 2001;105:115–126. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

