NAC1 regulates the recruitment of the proteasome complex into dendritic spines
- PMID: 17699672
- PMCID: PMC6672176
- DOI: 10.1523/JNEUROSCI.1571-07.2007
NAC1 regulates the recruitment of the proteasome complex into dendritic spines
Abstract
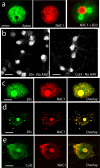
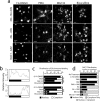
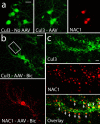
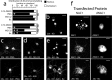
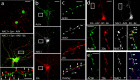
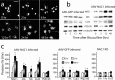
Coordinated proteolysis of synaptic proteins is required for synaptic plasticity, but a mechanism for recruiting the ubiquitin-proteasome system (UPS) into dendritic spines is not known. NAC1 is a cocaine-regulated transcriptional protein that was found to complex with proteins in the UPS, including cullins and Mov34. NAC1 and the proteasome were cotranslocated from the nucleus into dendritic spines in cortical neurons in response to proteasome inhibition or disinhibiting synaptic activity with bicuculline. Bicuculline also produced a progressive accumulation of the proteasome and NAC1 in the postsynaptic density. Recruitment of the proteasome into dendrites and postsynaptic density by bicuculline was prevented in neurons from mice harboring an NAC1 gene deletion or in neurons transfected with mutated NAC1 lacking the proteasome binding domain. These experiments show that NAC1 modulates the translocation of the UPS from the nucleus into dendritic spines, thereby suggesting a potential missing link in the recruitment of necessary proteolysis machinery for synaptic remodeling.
Figures

Similar articles
-
Growth-associated protein-43 is degraded via the ubiquitin-proteasome system.J Neurosci Res. 2005 Mar 1;79(5):652-60. doi: 10.1002/jnr.20388. J Neurosci Res. 2005. PMID: 15668958
-
NAC1, A POZ/BTB protein interacts with Parkin and may contribute to Parkinson's disease.Neuroscience. 2014 Jan 17;257:86-95. doi: 10.1016/j.neuroscience.2013.11.001. Epub 2013 Nov 11. Neuroscience. 2014. PMID: 24231739
-
The ubiquitin-proteasome system functionally links neuronal Tomosyn-1 to dendritic morphology.J Biol Chem. 2018 Feb 16;293(7):2232-2246. doi: 10.1074/jbc.M117.815514. Epub 2017 Dec 21. J Biol Chem. 2018. PMID: 29269412 Free PMC article.
-
Role of the ubiquitin-proteasome system in brain ischemia: friend or foe?Prog Neurobiol. 2014 Jan;112:50-69. doi: 10.1016/j.pneurobio.2013.10.003. Epub 2013 Oct 22. Prog Neurobiol. 2014. PMID: 24157661 Review.
-
The ubiquitin-proteasome system and learning-dependent synaptic plasticity - A 10 year update.Neurosci Biobehav Rev. 2023 Sep;152:105280. doi: 10.1016/j.neubiorev.2023.105280. Epub 2023 Jun 12. Neurosci Biobehav Rev. 2023. PMID: 37315660 Free PMC article. Review.
Cited by
-
An intellectual-disability-associated mutation of the transcriptional regulator NACC1 impairs glutamatergic neurotransmission.Front Mol Neurosci. 2023 Jul 14;16:1115880. doi: 10.3389/fnmol.2023.1115880. eCollection 2023. Front Mol Neurosci. 2023. PMID: 37533751 Free PMC article. Review.
-
Regulation of the proteasome by neuronal activity and calcium/calmodulin-dependent protein kinase II.J Biol Chem. 2009 Sep 25;284(39):26655-65. doi: 10.1074/jbc.M109.021956. Epub 2009 Jul 28. J Biol Chem. 2009. PMID: 19638347 Free PMC article.
-
Nacc1 Mutation in Mice Models Rare Neurodevelopmental Disorder with Underlying Synaptic Dysfunction.J Neurosci. 2024 Apr 3;44(14):e1610232024. doi: 10.1523/JNEUROSCI.1610-23.2024. J Neurosci. 2024. PMID: 38388424 Free PMC article.
-
A New Case of de novo Variant c.892C>T (p.Arg298Trp) in NACC1: A First Case Report From China.Front Pediatr. 2021 Nov 15;9:754261. doi: 10.3389/fped.2021.754261. eCollection 2021. Front Pediatr. 2021. PMID: 34869110 Free PMC article.
-
MicroRNAs signatures as potential molecular markers in mild cognitive impairment: a meta-analysis.Front Aging Neurosci. 2025 Jan 15;16:1524622. doi: 10.3389/fnagi.2024.1524622. eCollection 2024. Front Aging Neurosci. 2025. PMID: 39881680 Free PMC article.
References
-
- Allen E, Ding J, Wang W, Pramanik S, Chou J, Yau V, Yang Y. Gigaxonin-controlled degradation of MAP1B light chain is critical to neuronal survival. Nature. 2005;438:224–228. - PubMed
-
- Bingol B, Schuman EM. Synaptic protein degradation by the ubiquitin proteasome system. Curr Opin Neurobiol. 2005;15:536–541. - PubMed
-
- Bingol B, Schuman EM. Activity-dependent dynamics and sequestration of proteasomes in dendritic spines. Nature. 2006;441:144–1148. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
