Environmental signal integration by a modular AND gate
- PMID: 17700541
- PMCID: PMC1964800
- DOI: 10.1038/msb4100173
Environmental signal integration by a modular AND gate
Abstract
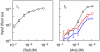
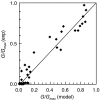
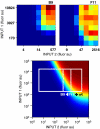
Microorganisms use genetic circuits to integrate environmental information. We have constructed a synthetic AND gate in the bacterium Escherichia coli that integrates information from two promoters as inputs and activates a promoter output only when both input promoters are transcriptionally active. The integration occurs via an interaction between an mRNA and tRNA. The first promoter controls the transcription of a T7 RNA polymerase gene with two internal amber stop codons blocking translation. The second promoter controls the amber suppressor tRNA supD. When both components are transcribed, T7 RNA polymerase is synthesized and this in turn activates a T7 promoter. Because inputs and outputs are promoters, the design is modular; that is, it can be reconnected to integrate different input signals and the output can be used to drive different cellular responses. We demonstrate this modularity by wiring the gate to integrate natural promoters (responding to Mg(2+) and AI-1) and using it to implement a phenotypic output (invasion of mammalian cells). A mathematical model of the transfer function is derived and parameterized using experimental data.
Figures



References
-
- Anderson JC, Clarke EJ, Arkin AP, Voigt CA (2006) Environmentally controlled invasion of cancer cells by engineered bacteria. J Mol Biol 355: 619–627 - PubMed
-
- Anderson JC, Magliery TJ, Schultz PG (2002) Exploring the limits of codon and anticodon size. Chem Biol 9: 237–244 - PubMed
-
- Bassler BL, Losick R (2006) Bacterially speaking. Cell 125: 237–246 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

