Classification of human chromosome 21 gene-expression variations in Down syndrome: impact on disease phenotypes
- PMID: 17701894
- PMCID: PMC1950826
- DOI: 10.1086/520000
Classification of human chromosome 21 gene-expression variations in Down syndrome: impact on disease phenotypes
Abstract
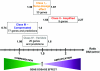
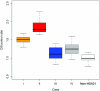
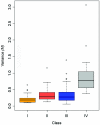
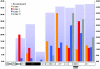
Down syndrome caused by chromosome 21 trisomy is the most common genetic cause of mental retardation in humans. Disruption of the phenotype is thought to be the result of gene-dosage imbalance. Variations in chromosome 21 gene expression in Down syndrome were analyzed in lymphoblastoid cells derived from patients and control individuals. Of the 359 genes and predictions displayed on a specifically designed high-content chromosome 21 microarray, one-third were expressed in lymphoblastoid cells. We performed a mixed-model analysis of variance to find genes that are differentially expressed in Down syndrome independent of sex and interindividual variations. In addition, we identified genes with variations between Down syndrome and control samples that were significantly different from the gene-dosage effect (1.5). Microarray data were validated by quantitative polymerase chain reaction. We found that 29% of the expressed chromosome 21 transcripts are overexpressed in Down syndrome and correspond to either genes or open reading frames. Among these, 22% are increased proportional to the gene-dosage effect, and 7% are amplified. The other 71% of expressed sequences are either compensated (56%, with a large proportion of predicted genes and antisense transcripts) or highly variable among individuals (15%). Thus, most of the chromosome 21 transcripts are compensated for the gene-dosage effect. Overexpressed genes are likely to be involved in the Down syndrome phenotype, in contrast to the compensated genes. Highly variable genes could account for phenotypic variations observed in patients. Finally, we show that alternative transcripts belonging to the same gene are similarly regulated in Down syndrome but sense and antisense transcripts are not.
Figures

References
Web Resources
-
- Eleanor Roosevelt Institute: Chromosome 21 Gene Function and Pathway Database, http://chr21db.cudenver.edu/
-
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for accession numbers in tables 5–8)
-
- Gene Expression Omnibus (GEO), http://www.ncbi.nlm.nih.gov/geo/ (for accession number GSE6408)
-
- Max Planck Institute: Chromosome 21 Gene Catalog Based on the New AGP File July 2002, http://chr21.molgen.mpg.de/chr21_catalogs/chr21_mar_2002.html
-
- NCBI Entrez, http://www.ncbi.nlm.nih.gov/gquery/gquery.fcgi (for accession numbers L13852 and AB000468)
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

