Evidence of still-ongoing convergence evolution of the lactase persistence T-13910 alleles in humans
- PMID: 17701907
- PMCID: PMC1950831
- DOI: 10.1086/520705
Evidence of still-ongoing convergence evolution of the lactase persistence T-13910 alleles in humans
Abstract
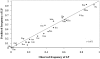
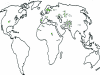
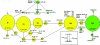
A single-nucleotide variant, C/T(-13910), located 14 kb upstream of the lactase gene (LCT), has been shown to be completely correlated with lactase persistence (LP) in northern Europeans. Here, we analyzed the background of the alleles carrying the critical variant in 1,611 DNA samples from 37 populations. Our data show that the T(-13910) variant is found on two different, highly divergent haplotype backgrounds in the global populations. The first is the most common LP haplotype (LP H98) present in all populations analyzed, whereas the others (LP H8-H12), which originate from the same ancestral allelic haplotype, are found in geographically restricted populations living west of the Urals and north of the Caucasus. The global distribution pattern of LP T(-13910) H98 supports the Caucasian origin of this allele. Age estimates based on different mathematical models show that the common LP T(-13910) H98 allele (approximately 5,000-12,000 years old) is relatively older than the other geographically restricted LP alleles (approximately 1,400-3,000 years old). Our data about global allelic haplotypes of the lactose-tolerance variant imply that the T(-13910) allele has been independently introduced more than once and that there is a still-ongoing process of convergent evolution of the LP alleles in humans.
Figures
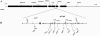
Similar articles
-
Independent introduction of two lactase-persistence alleles into human populations reflects different history of adaptation to milk culture.Am J Hum Genet. 2008 Jan;82(1):57-72. doi: 10.1016/j.ajhg.2007.09.012. Am J Hum Genet. 2008. PMID: 18179885 Free PMC article.
-
Several different lactase persistence associated alleles and high diversity of the lactase gene in the admixed Brazilian population.PLoS One. 2012;7(9):e46520. doi: 10.1371/journal.pone.0046520. Epub 2012 Sep 28. PLoS One. 2012. PMID: 23029545 Free PMC article.
-
Lactose digestion and the evolutionary genetics of lactase persistence.Hum Genet. 2009 Jan;124(6):579-91. doi: 10.1007/s00439-008-0593-6. Epub 2008 Nov 26. Hum Genet. 2009. PMID: 19034520
-
From 'lactose intolerance' to 'lactose nutrition'.Asia Pac J Clin Nutr. 2015;24 Suppl 1:S1-8. doi: 10.6133/apjcn.2015.24.s1.01. Asia Pac J Clin Nutr. 2015. PMID: 26715078 Review.
-
Genetics of lactase persistence and lactose intolerance.Annu Rev Genet. 2003;37:197-219. doi: 10.1146/annurev.genet.37.110801.143820. Annu Rev Genet. 2003. PMID: 14616060 Review.
Cited by
-
Isolated populations and complex disease gene identification.Genome Biol. 2008;9(8):109. doi: 10.1186/gb-2008-9-8-109. Epub 2008 Aug 26. Genome Biol. 2008. PMID: 18771588 Free PMC article.
-
Update on lactose malabsorption and intolerance: pathogenesis, diagnosis and clinical management.Gut. 2019 Nov;68(11):2080-2091. doi: 10.1136/gutjnl-2019-318404. Epub 2019 Aug 19. Gut. 2019. PMID: 31427404 Free PMC article. Review.
-
Independent introduction of two lactase-persistence alleles into human populations reflects different history of adaptation to milk culture.Am J Hum Genet. 2008 Jan;82(1):57-72. doi: 10.1016/j.ajhg.2007.09.012. Am J Hum Genet. 2008. PMID: 18179885 Free PMC article.
-
Dairy Intake and Body Composition and Cardiometabolic Traits among Adults: Mendelian Randomization Analysis of 182041 Individuals from 18 Studies.Clin Chem. 2019 Jun;65(6):751-760. doi: 10.1373/clinchem.2018.300335. Clin Chem. 2019. PMID: 31138550 Free PMC article.
-
Evolution of Lactase Persistence: Turbo-Charging Adaptation in Growth Under the Selective Pressure of Maternal Mortality?Front Physiol. 2021 Aug 23;12:696516. doi: 10.3389/fphys.2021.696516. eCollection 2021. Front Physiol. 2021. PMID: 34497534 Free PMC article.
References
Web Resources
-
- Arlequin, http://lgb.unige.ch/arlequin/
-
- dbSNP, http://www.ncbi.nlm.nih.gov/SNP/ (for SNPs 2 [rs3754686], 3 [rs3769005], 4 [rs4988235], 5 [rs4954493], 6 [rs3099181]), 7 [rs182549], 8 [rs4988183], and 9 [rs3087343])
-
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for indel polymorphism sequence within intron 1 of LCT [accession number DQ109677])
-
- NETWORK version 4.1.1.2, http://www.fluxus-engineering.com/
-
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for lactase, LNP, and LP)
References
-
- Sahi T (1994) Genetics and epidemiology of adult-type hypolactasia. Scand J Gastroenterol Suppl 202:7–20 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

