Crystal structure of an ancient protein: evolution by conformational epistasis
- PMID: 17702911
- PMCID: PMC2519897
- DOI: 10.1126/science.1142819
Crystal structure of an ancient protein: evolution by conformational epistasis
Abstract
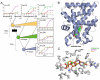
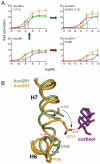
The structural mechanisms by which proteins have evolved new functions are known only indirectly. We report x-ray crystal structures of a resurrected ancestral protein-the approximately 450 million-year-old precursor of vertebrate glucocorticoid (GR) and mineralocorticoid (MR) receptors. Using structural, phylogenetic, and functional analysis, we identify the specific set of historical mutations that recapitulate the evolution of GR's hormone specificity from an MR-like ancestor. These substitutions repositioned crucial residues to create new receptor-ligand and intraprotein contacts. Strong epistatic interactions occur because one substitution changes the conformational position of another site. "Permissive" mutations-substitutions of no immediate consequence, which stabilize specific elements of the protein and allow it to tolerate subsequent function-switching changes-played a major role in determining GR's evolutionary trajectory.
Figures


References
-
- Golding GB, Dean AM. Mol Biol Evol. 1998;15:355. - PubMed
-
- Stern DL. Evolution Int J Org Evolution. 2000;54:1079. - PubMed
-
- Perutz MF. Mol Biol Evol. 1983;1:1. - PubMed
-
- Glasner ME, Gerlt JA, Babbitt PC. Curr Opin Chem Biol. 2006;10:492. - PubMed
-
- Kinch LN, Grishin NV. Curr Opin Struct Biol. 2002;12:400. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

