Comprehensive testing of positionally cloned asthma genes in two populations
- PMID: 17702965
- PMCID: PMC2048676
- DOI: 10.1164/rccm.200704-592OC
Comprehensive testing of positionally cloned asthma genes in two populations
Abstract
Rationale: Replication of gene-disease associations has become a requirement in complex trait genetics.
Objectives: In studies of childhood asthma from two different ethnic groups, we attempted to replicate associations with five potential asthma susceptibility genes previously identified by positional cloning.
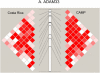
Methods: We analyzed two family-based samples ascertained through an asthmatic proband: 497 European-American children from the Childhood Asthma Management Program and 439 Hispanic children from the Central Valley of Costa Rica. We genotyped 98 linkage disequilibrium-tagging single-nucleotide polymorphisms (SNPs) in five genes: ADAM33, DPP10, GPR154 (HUGO name: NPSR1), HLA-G, and the PHF11 locus (includes genes SETDB2 and RCBTB1). SNPs were tested for association with asthma and two intermediate phenotypes: airway hyperresponsiveness and total serum immunoglobulin E levels.
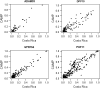
Measurements and main results: Despite differing ancestries, linkage disequilibrium patterns were similar in both cohorts. Of the five evaluated genes, SNP-level replication was found only for GPR154 (NPSR1). In this gene, three SNPs were associated with asthma in both cohorts, although the opposite alleles were associated in either study. Weak evidence for locus-level replication with asthma was found in the PHF11 locus, although there was no overlap in the associated SNP across the two cohorts. No consistent associations were observed for the three other genes.
Conclusions: These results provide some further support for the role of genetic variation in GPR154 (NPSR1) and PHF11 in asthma susceptibility and also highlight the challenges of replicating genetic associations in complex traits such as asthma, even for genes identified by linkage analysis.
Figures



Similar articles
-
Polymorphisms of PHF11 and DPP10 are associated with asthma and related traits in a Chinese population.Respiration. 2010;79(1):17-24. doi: 10.1159/000235545. Epub 2009 Aug 11. Respiration. 2010. PMID: 19672052
-
Positionally cloned asthma susceptibility gene polymorphisms and disease risk in the British 1958 Birth Cohort.Thorax. 2009 May;64(5):381-7. doi: 10.1136/thx.2008.102053. Epub 2009 Feb 22. Thorax. 2009. PMID: 19237393
-
Analyses of associations between three positionally cloned asthma candidate genes and asthma or asthma-related phenotypes in a Chinese population.BMC Med Genet. 2009 Dec 1;10:123. doi: 10.1186/1471-2350-10-123. BMC Med Genet. 2009. PMID: 19951440 Free PMC article.
-
Asthma genetics 2003.Hum Mol Genet. 2004 Apr 1;13 Spec No 1:R83-9. doi: 10.1093/hmg/ddh080. Epub 2004 Feb 5. Hum Mol Genet. 2004. PMID: 14764626 Review.
-
Mapping and identifying genes for asthma and psoriasis.Philos Trans R Soc Lond B Biol Sci. 2005 Aug 29;360(1460):1551-61. doi: 10.1098/rstb.2005.1684. Philos Trans R Soc Lond B Biol Sci. 2005. PMID: 16096103 Free PMC article. Review.
Cited by
-
Thymic stromal lymphopoietin (TSLP) is associated with allergic rhinitis in children with asthma.Clin Mol Allergy. 2011 Jan 18;9:1. doi: 10.1186/1476-7961-9-1. Clin Mol Allergy. 2011. PMID: 21244681 Free PMC article.
-
Genome-wide analysis of survival in early-stage non-small-cell lung cancer.J Clin Oncol. 2009 Jun 1;27(16):2660-7. doi: 10.1200/JCO.2008.18.7906. Epub 2009 May 4. J Clin Oncol. 2009. PMID: 19414679 Free PMC article.
-
Genetic variations in GRIA1 on chromosome 5q33 related to asparaginase hypersensitivity.Clin Pharmacol Ther. 2010 Aug;88(2):191-6. doi: 10.1038/clpt.2010.94. Epub 2010 Jun 30. Clin Pharmacol Ther. 2010. PMID: 20592726 Free PMC article.
-
Novel eosinophilic gene expression networks associated with IgE in two distinct asthma populations.Clin Exp Allergy. 2018 Dec;48(12):1654-1664. doi: 10.1111/cea.13249. Epub 2018 Nov 21. Clin Exp Allergy. 2018. PMID: 30107053 Free PMC article.
-
Race, ethnicity and social class and the complex etiologies of asthma.Pharmacogenomics. 2008 Apr;9(4):453-62. doi: 10.2217/14622416.9.4.453. Pharmacogenomics. 2008. PMID: 18384258 Free PMC article. Review.
References
-
- Freely associating [editorial]. Nat Genet 1999;22:1–2. - PubMed
-
- Hirschhorn JN, Altshuler D. Once and again: issues surrounding replication in genetic association studies. J Clin Endocrinol Metab 2002;87:4438–4441. - PubMed
-
- Ioannidis JP, Ntzani EE, Trikalinos TA, Contopoulos-Ioannidis DG. Replication validity of genetic association studies. Nat Genet 2001;29:306–309. - PubMed
-
- Lemanske RF Jr, Busse WW. Asthma. JAMA 1997;278:1855–1873. - PubMed
-
- Bloom B, Dey AN. Summary health statistics for US children: National Health Interview Survey, 2004. Vital Health Stat 10 2006;227:1–85. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

