Coarse-grained free energy functions for studying protein conformational changes: a double-well network model
- PMID: 17704151
- PMCID: PMC2084241
- DOI: 10.1529/biophysj.107.112060
Coarse-grained free energy functions for studying protein conformational changes: a double-well network model
Abstract
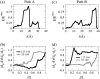
In this work, a double-well network model (DWNM) is presented for generating a coarse-grained free energy function that can be used to study the transition between reference conformational states of a protein molecule. Compared to earlier work that uses a single, multidimensional double-well potential to connect two conformational states, the DWNM uses a set of interconnected double-well potentials for this purpose. The DWNM free energy function has multiple intermediate states and saddle points, and is hence a "rough" free energy landscape. In this implementation of the DWNM, the free energy function is reduced to an elastic-network model representation near the two reference states. The effects of free energy function roughness on the reaction pathways of protein conformational change is demonstrated by applying the DWNM to the conformational changes of two protein systems: the coil-to-helix transition of the DB-loop in G-actin and the open-to-closed transition of adenylate kinase. In both systems, the rough free energy function of the DWNM leads to the identification of distinct minimum free energy paths connecting two conformational states. These results indicate that while the elastic-network model captures the low-frequency vibrational motions of a protein, the roughness in the free energy function introduced by the DWNM can be used to characterize the transition mechanism between protein conformations.
Figures


References
-
- Lodish, H., M. P. Scott, P. Matsudaira, J. Darnell, L. Zipursky, C. A. Kaiser, A. Berk, and M. Krieger. 2003. Molecular Cell Biology. W. H. Freeman, New York.
-
- Ackers, G. K. 1998. Deciphering the molecular code of hemoglobin allostery. Adv. Protein Chem. 51:185–253. - PubMed
-
- Crivici, A., and M. Ikura. 1995. Molecular and structural basis of target recognition by calmodulin. Annu. Rev. Biophys. Biomol. Struct. 24:85–116. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

