How dopamine transporter interacts with dopamine: insights from molecular modeling and simulation
- PMID: 17704152
- PMCID: PMC2072054
- DOI: 10.1529/biophysj.107.110924
How dopamine transporter interacts with dopamine: insights from molecular modeling and simulation
Abstract
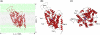
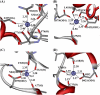
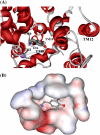
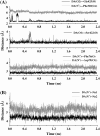
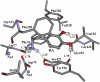
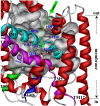
By performing homology modeling, molecular docking, and molecular dynamics simulations, we have developed three-dimensional (3D) structural models of both dopamine transporter and dopamine transporter-dopamine complex in the environment of lipid bilayer and solvent water. According to the simulated structure of dopamine transporter-dopamine complex, dopamine was orientated in a hydrophobic pocket at the midpoint of the membrane. The modeled 3D structures provide some detailed structural and mechanistic insights concerning how dopamine transporter (DAT) interacts with dopamine at atomic level, extending our mechanistic understanding of the dopamine reuptake with the help of Na(+) ions. The general features of the modeled 3D structures are consistent with available experimental data. Based on the modeled structures, our calculated binding free energy (DeltaG(bind) = -6.4 kcal/mol) for dopamine binding with DAT is also reasonably close to the experimentally derived DeltaG(bind) value of -7.4 kcal/mol. Finally, a possible dopamine-entry pathway, which involves formation and breaking of the salt bridge between side chains of Arg(85) and Asp(476), is proposed based on the results obtained from the modeling and molecular dynamics simulation. The new structural and mechanistic insights obtained from this computational study are expected to stimulate future, further biochemical and pharmacological studies on the detailed structures and mechanisms of DAT and other homologous transporters.
Figures


Similar articles
-
Insights from molecular dynamics: the binding site of cocaine in the dopamine transporter and permeation pathways of substrates in the leucine and dopamine transporters.J Mol Graph Model. 2012 Sep;38:1-12. doi: 10.1016/j.jmgm.2012.05.007. Epub 2012 Jun 19. J Mol Graph Model. 2012. PMID: 23079638 Free PMC article.
-
Molecular Mechanism of Dopamine Transport by Human Dopamine Transporter.Structure. 2015 Nov 3;23(11):2171-81. doi: 10.1016/j.str.2015.09.001. Epub 2015 Oct 15. Structure. 2015. PMID: 26481814 Free PMC article.
-
Molecular model of the neural dopamine transporter.J Comput Aided Mol Des. 2003 May-Jun;17(5-6):367-82. doi: 10.1023/a:1026116017725. J Comput Aided Mol Des. 2003. PMID: 14635728
-
Emerging structure-function relationships defining monoamine NSS transporter substrate and ligand affinity.Biochem Pharmacol. 2010 Apr 15;79(8):1083-91. doi: 10.1016/j.bcp.2009.11.019. Epub 2009 Nov 30. Biochem Pharmacol. 2010. PMID: 19954741 Review.
-
A comprehensive atlas of the topography of functional groups of the dopamine transporter.Synapse. 2005 Nov;58(2):72-94. doi: 10.1002/syn.20183. Synapse. 2005. PMID: 16088952 Review.
Cited by
-
The atypical stimulant and nootropic modafinil interacts with the dopamine transporter in a different manner than classical cocaine-like inhibitors.PLoS One. 2011;6(10):e25790. doi: 10.1371/journal.pone.0025790. Epub 2011 Oct 17. PLoS One. 2011. PMID: 22043293 Free PMC article.
-
Steric parameters, molecular modeling and hydropathic interaction analysis of the pharmacology of para-substituted methcathinone analogues.Br J Pharmacol. 2015 May;172(9):2210-8. doi: 10.1111/bph.13043. Epub 2015 Feb 27. Br J Pharmacol. 2015. PMID: 25522019 Free PMC article.
-
Modeling binding modes of alpha7 nicotinic acetylcholine receptor with ligands: the roles of Gln117 and other residues of the receptor in agonist binding.J Med Chem. 2008 Oct 23;51(20):6293-302. doi: 10.1021/jm800607u. Epub 2008 Oct 1. J Med Chem. 2008. PMID: 18826295 Free PMC article.
-
In Silico Investigations into the Selectivity of Psychoactive and New Psychoactive Substances in Monoamine Transporters.ACS Omega. 2022 Oct 21;7(43):38311-38321. doi: 10.1021/acsomega.2c02714. eCollection 2022 Nov 1. ACS Omega. 2022. PMID: 36340072 Free PMC article.
-
Binding Mode of Human Norepinephrine Transporter Interacting with HIV-1 Tat.ACS Chem Neurosci. 2021 May 5;12(9):1519-1527. doi: 10.1021/acschemneuro.0c00792. Epub 2021 Apr 22. ACS Chem Neurosci. 2021. PMID: 33886267 Free PMC article.
References
-
- Uhl, R. G. 2003. Dopamine transporters: basic science and human variation of a key molecule for dopaminergic function, locomotion, and Parkinsonism. Mov. Disord. 18:S71–S80. - PubMed
-
- Mendelson, J. H., and N. K. Mello. 1996. Management of cocaine abuse and dependence. N. Engl. J. Med. 334:965–972. - PubMed
-
- Singh, S. 2000. Chemistry, design, and structure-activity relationship of cocaine antagonists. Chem. Rev. 100:925–1024. - PubMed
-
- Paula, S., M. R. Tabet, C. D. Farr, A. B. Norman, and W. J. Jr. Ball. 2004. Three-dimensional quantitative structure-activity relationship modeling of cocaine binding by a novel human monoclonal antibody. J. Med. Chem. 47:133–142. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

