Effects of heavy fuel oil on the bacterial community structure of a pristine microbial mat
- PMID: 17704271
- PMCID: PMC2075027
- DOI: 10.1128/AEM.01352-07
Effects of heavy fuel oil on the bacterial community structure of a pristine microbial mat
Abstract
The effects of petroleum contamination on the bacterial community of a pristine microbial mat from Salins-de-Giraud (Camargue, France) have been investigated. Mats were maintained as microcosms and contaminated with no. 2 fuel oil from the wreck of the Erika. The evolution of the complex bacterial community was monitored by combining analyses based on 16S rRNA genes and their transcripts. 16S rRNA gene-based terminal restriction fragment length polymorphism (T-RFLP) analyses clearly showed the effects of the heavy fuel oil after 60 days of incubation. At the end of the experiment, the initial community structure was recovered, illustrating the resilience of this microbial ecosystem. In addition, the responses of the metabolically active bacterial community were evaluated by T-RFLP and clone library analyses based on 16S rRNA. Immediately after the heavy fuel oil was added to the microcosms, the structure of the active bacterial community was modified, indicating a rapid microbial mat response. Members of the Gammaproteobacteria were initially dominant in the contaminated microcosms. Pseudomonas and Acinetobacter were the main genera representative of this class. After 90 days of incubation, the Gammaproteobacteria were superseded by "Bacilli" and Alphaproteobacteria. This study shows the major changes that occur in the microbial mat community at different time periods following contamination. At the conclusion of the experiment, the RNA approach also demonstrated the resilience of the microbial mat community in resisting environmental stress resulting from oil pollution.
Figures
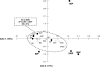
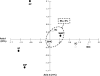


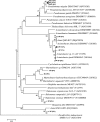
References
-
- Abed, R. M. M., and J. Köster. 2005. The direct role of aerobic heterotrophic bacteria associated with cyanobacteria in the degradation of oil compounds. Int. Biodeterior. Biodegradation 55:29-37.
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
-
- Bender, J., and P. Phillips. 2004. Microbial mats for multiple applications in aquaculture and bioremediation. Bioresour. Technol. 94:229-238. - PubMed
-
- Berge, J. A., R. G. Lichtenthaler, and F. Oreld. 1987. Hydrocarbon depuration and abiotic changes in artificially oil contaminated sediment in the subtidal. Estuar. Coast. Shelf Sci. 24:567-568.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

